* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download AP Biology Discussion Notes
Survey
Document related concepts
Zinc finger nuclease wikipedia , lookup
DNA sequencing wikipedia , lookup
DNA repair protein XRCC4 wikipedia , lookup
DNA profiling wikipedia , lookup
Homologous recombination wikipedia , lookup
DNA nanotechnology wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Microsatellite wikipedia , lookup
Eukaryotic DNA replication wikipedia , lookup
DNA polymerase wikipedia , lookup
Helitron (biology) wikipedia , lookup
Transcript
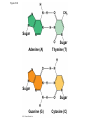
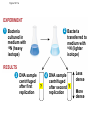
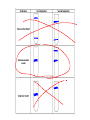
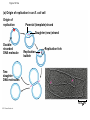
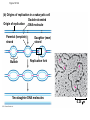
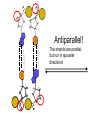
AP Biology Discussion Notes Thursday 01/26/2017 Goals for Today • Be able to say what the central dogma of biology is. • Be able to describe the structure of DNA and its associated terms • Be able to describe the process of DNA replication & Terms involved • Be able to “build” a strand of DNA 1/26 Question of the Day • List the 2 groups of Nitrogen bases and which bases belong to each group. Then star the group with the larger (2-ring) bases. (Hint we are not talking about pairing.) *Bases always pair Pyrimidine to _________ Review – Discuss with a partner 1. Identify and describe what 2 things prevent “mismatches” in DNA base pairing. Figure 16.UN01 Purine purine: too wide Pyrimidine pyrimidine: too narrow Purine pyrimidine: width consistent with X-ray data Figure 16.8 Sugar Sugar Adenine (A) Thymine (T) Sugar Sugar Guanine (G) Cytosine (C) Review – Discuss with a partner 1. Explain what is meant by the term “DNA replication” 2. Identify what term describe this process and explain what that means. (Is it liberal?) Figure 16.9-1 Semiconservative Replication A T C G T A A T G C (a) Parent molecule Figure 16.9-2 Semiconservative Replication A T A T C G C G T A T A A T A T G C G C (a) Parent molecule (b) Separation of strands Figure 16.9-3 Semiconservative Replication A T A T A T A T C G C G C G C G T A T A T A T A A T A T A T A T G C G C G C G C (a) Parent molecule (b) Separation of strands (c) “Daughter” DNA molecules, each consisting of one parental strand and one new strand Chapter 16 in your book Figure 16.10 (a) Conservative model (b) Semiconservative model (c) Dispersive model Parent cell First replication Second replication • Experiments by Matthew Meselson and Franklin Stahl supported the semiconservative model • They labeled the nucleotides of the old strands with a heavy isotope of nitrogen 15N, while any new nucleotides were labeled with a lighter isotope, 14N Figure 16.11a EXPERIMENT 2 Bacteria transferred to medium with 14N (lighter isotope) 1 Bacteria cultured in medium with 15N (heavy isotope) RESULTS 3 DNA sample centrifuged after first replication ? 4 DNA sample centrifuged after second ? replication Less dense More dense Figure 16.11b CONCLUSION Predictions: First replication Conservative model Semiconservative model Dispersive model Second replication DNA Replication: A Closer Look • The copying of DNA is remarkable in its speed and accuracy –E.coli has about 4.6 Million nucleotide/base pairs and replicates DNA, then divides into 2 new cells in less than an hour! –Humans have ~6 Billion nucleotide/base pairs and replicate their DNA in a few hours Getting Started • Replication begins at particular sites called origins of replication, where the two DNA strands are separated, opening up a replication “bubble” • A eukaryotic chromosome may have hundreds or even thousands of origins of replication • Replication proceeds in both directions from each origin, until the entire* molecule is copied Figure 16.12a (a) Origin of replication in an E. coli cell Origin of replication Parental (template) strand Daughter (new) strand Doublestranded DNA molecule Replication bubble Replication fork Two daughter DNA molecules 0.5 m Synthesizing a New DNA Strand • Enzymes called DNA polymerases catalyze the elongation of new DNA at a replication fork • The rate of elongation is about 500 nucleotides per second in bacteria and 50 per second in human cells • How do we speed this up in our cells? Figure 16.12b (b) Origins of replication in a eukaryotic cell Double-stranded Origin of replication DNA molecule Parental (template) strand Bubble Daughter (new) strand Replication fork Two daughter DNA molecules 0.25 m DNA Replication: Cast of Characters • The Enzymes* – Helicase (“Hacks”) – DNA Polymerase (“Pastes”) • READS: • BUILDS: – Ligase (“Links”) Polymerase only builds in 5’ to 3’ Direction! Remember: it’s an enzyme - & they’re very picky Antiparallel? Antiparallel! The strands are parallel, but run in opposite directions! DNA Replication: Cast of Characters • Building Blocks: – Nucleotides (A,T,C,G) • Strand Terminology: – Origin(s) of Replication – Replication Fork – Leading Strand – Lagging Strand • Okazaki Fragments • DNA Replication in Action Questions? DNA Assignment • DUE – Wednesday– February 1st!