* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 1 - KOCW
List of types of proteins wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transcription factor wikipedia , lookup
Molecular evolution wikipedia , lookup
Maurice Wilkins wikipedia , lookup
Gene expression wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Molecular cloning wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
DNA supercoil wikipedia , lookup
Acetylation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
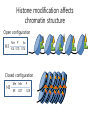
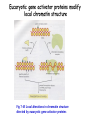
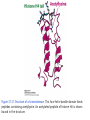
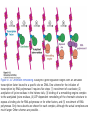
Histone modifications Nucleosomes Are Complexes of DNA and Histones Eukaryotic DNA is not naked; chromatin, DNA plus histones (50%) Five major histones; H2A, H2B, H3, H4 and H1 highly basic (Arg and Lys >25%) Roger Kornberg in 1974; Chromatin is made up of repeating units called nucleosomes, each containing 200 bp of DNA and two copies each of H2A, H2B, H3, H4 (histone octamer); beads on a string, nucleosome core particle plus linker DNA (H1 binds to this DNA) Figure 31.19 Chromatin structure. An electron micrograph of chromatin showing its “beads on a string” character. Nucleosome core particle (~100Å in diameter); ~145 bp DNA wrapped around histone octamer Figure 31.20 Nucleosome core particle. The structure consists of a core of eight histone proteins surrounded by DNA. (A) A view showing the DNA wrapping around the histone core. (B) A view related to that in part A by a 90-degree rotation. Notice that the DNA forms a left-handed superhelix as it wraps around the core. (C) A schematic view.. Eukaryotic DNA is packaged in nucleosomes Approximately 147 base pa irs of DNA wrapped around a histone octamer ~ 147 bp DNA + 8 histones: 2 each H2A H2B H3 H4 Eukaryotic DNA Is Wrapped Around Histones to Form Nucleosomes The four types of histone are homologous and similar in structure An histone octomer consists of a (H3)2(H4)2 tetramer and a pair of H2A-H2B dimers to form a left-handed superhelical ramp around which the DNA wraps. Each histone has an flexible N-terminal tail rich in Lys and Arg that extends out from the core structure. Covalent modification of these tails play an essential role in modulating the affinity of the histones for DNA and other proteins. Figure 31.21 Homologous histones. Histones H2A, H2B, H3, and H4 adopt a similar three-dimensional structure as a consequence of common ancestry. Some parts of the tails at the termini of the proteins are not shown. Chromatin Structure Is Modulated Through Covalent Modifications of Histone Tails How do the coactivator modulate transcription activity? Some p160 coactivators are histone acetyltransferases (HATs); transfer acetyl group from acetyl CoA to the e amino group of specific lysine residues in the N-terminal tail of histones. 1. Changes in affinity to DNA; the acetylated histone tail or the entire histone complex 2. Recruitment of the bromodomain (acetyllysine-binding domain) proteins a. TAFs (TATA-box binding protein (TBP) associated factors). TAF1 contains two bromodomains for positions 5 and 12 in the H4 tail b. the chromatin remodeling factor to reorganize chromatin structure Figure 31.30 Structure of histone acetyltransferase. The amino-terminal tail of histone H3 extends into a pocket in which a lysine side chain can acceptor an acetyl group from acetyl CoA bound in an adjacent site. Histone proteins can be modified to affect chromatin structure DNA Histone octamer NUCLEOSOME The amino terminal regions of the histone monomers ex tend beyond the nucleosom e and are accessible for mo dification. The Histone Code • Histones can be modified by – – – – – Acetylation (Ac) Ubiquitination (Ub) Methylation (Me) Phosphorylation (P) Sumoylation (Su) • Depending on their position, these can contribute to transcriptional activation or inactivation. Example – H3 modifications Me H3 Me P Ac Me Ac Ac Me Me P A R T K Q T A R K S T G G K A P R K Q L A T K A A R K S 4 9 10 14 1718 23 262728 The amino terminus of H3 is often modified at one or more positions, which can contribute to an activation or inhibition of transcription. Example – H3 modifications Me H3 Me P Ac Me Ac Ac Me Me P A R T K Q T A R K S T G G K A P R K Q L A T K A A R K S 4 9 10 14 1718 23 262728 The amino terminus of H3 is often modified at one or more positions, which can contribute to an activation or inhibition of transcription. Lysine can be acetylated, or mono-, di-, or tri-methylated Methylated lysine Mono (Kme1) + NH3 Lysine (K) N Di (Kme2) CH3 O Acetylated lysine (KAc) Tri (Kme3) + NH2 + N H + N CH3 CH3 CH3 CH3 CH3 CH3 Histone modification affects chromatin structure Open configuration H3 Me P Ac K4 S10 K14 Closed configuration H3 Me Me P K9 K27 S28 H3K27me3 is associated with genes BLUE = Gene density RED = Repetitive element density GREEN = H3K27me3 PURPLE = methylcytosine H3K27me3 in Arabidopsis is present within the generich region, not the repeat -rich region. epigenetic marks DNA demethylation DNA methylation Histone acetylation Histone deacetylation Histone (de) methylation Histone (de) methylation Histone variants Fig 7-48. An order of events leading to transcription initiation at a specific promoter Eucaryotic gene activator proteins modify local chromatin structure Fig 7-45 Local alterations in chromatin structure directed by eucaryotic gene activator proteins Fig 7-46 Two specific ways that local histone acetylation stimulate transcription initiation Figure 31.31 Structure of a bromodomain. This four-helix-bundle domain binds peptides containing acetyllysine. An acetylated peptide of histone H4 is shown bound in the structure. Figure 31.32 Chromatin remodeling. Eukaryotic gene regulation begins with an activated transcription factor bound to a specific site on DNA. One scheme for the initiation of transcription by RNA polymerase ll requires five steps: (1) recruitment of coactivator, (2) acetylation of lysine residues in the histone tails, (3) binding of a remodeling-engine complex to the acetylated lysine residues, (4) ATP-dependent remodeling of the chromatin structure to expose a binding site for RNA polymerase or for other factors, and (5) recruitment of RNA polymerase. Only two subunits are shown for each complex, although the actual complexes are much larger. Other schemes are possible. . Fig 7-49. (continued)