* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 3000_2013_2b
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene expression programming wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Medical genetics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Public health genomics wikipedia , lookup
Genome evolution wikipedia , lookup
Point mutation wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Behavioural genetics wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Koinophilia wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Designer baby wikipedia , lookup
Genome-wide association study wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Heritability of IQ wikipedia , lookup
Human genetic variation wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genetic drift wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Population genetics wikipedia , lookup
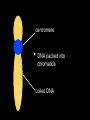
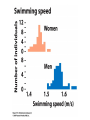
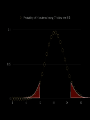
mutations are the basis of diversity we will learn THIS: [mutations happen and add diversity to a population. A/G, presence/absence, red/white] such diversity is temporary. in a finite population a single genetic polymorphism will eventually disappear through drift, but selection, non-random mating, and migration will modify that rate sciencedaily.com offspring = heredity • bacteria, archaea divide (literally) into two daughter cells • most eukaryotes make it more complicated: sexual reproduction centromere • DNA packed into chromatids coiled DNA • DNA packed into chromatids • sister chromatids contain homologous stretches of DNA and bound at centromere • each chromatid was inherited from different parent • ACTTCAGAT ACTTCAGAT • • GGTCATATAT GGTCACATAT homologous regions (LOCUS or LOCI) found on each chromatid are ALLELES they may be different at one or more nucleotides or other forms of mutation so: LOCUS descended from single ancestral region of genome; ALLELES represent the diversity of that locus so: each sexually produced offspring is unique because: mutations AND genetic recombination this diversity is what we are exploring! so: each sexually produced offspring is unique because: mutations AND genetic recombination this diversity is what we are exploring! diversity • diversity of GENOTYPE (combination of alleles at one or more LOCI) is one contribution to the organism PHENOTYPE (what it looks like, manifestation of genotype) • genes are inherited; phenotype is complex, involving genotype at one or more loci AND environmental contributions Gamete production Independent assortment Expected proportions and Combinations some phenotypes depend almost entirely on genotype at single locus (illustrated by Punnett squares) some phenotypes are not controlled by genotype ? normal distributions • “bell” curves, Gaussian, tend to have a mean (location of peak) and symmetric variance (the width of the distribution) • why do we see it so much? why does it apply to quantitative traits? • “central limit theorem”: sum of many random variables is distributed normally aa Aa AA 18 how do we find these many genes? • human body height, flower corolla length, many genes (quantitative) but where are they? • quantitative trait loci (QTL) • association studies allow us to find the markers that tend to be found in individuals with a given trait LOD: log (ratio) of odds, e.g. LOD of 2 means that a genetic marker is 2 10 or 100 times more likely associated with trait than not meaning, marker and trait show up together more often than expected by chance variation matters • dominant, recessive, homozygosity, heterozygosity, allelic variation of all types • important component of variation is the average effect alleles (e.g. having additiveof genetic variance symbolized as Var(A) allele GATGAT generates red pigment; variance in phenotype due to average effects of alleles GACGAT white pigment; heterozygotes are pink) • allele symbol discussion: A is not dominant to a (necessarily), just traditional symbols, ANY SYMBOL models • all models are wrong • some are useful • • science attempts to find the simplest set of interactions that explains the most complex observations “How do these genes combine to determine the phenotype of an individual? The simplest model is to assume that genes act additively with each other both within and between loci, but of course they may interact to show dominance or epistasis, respectively.” – Hill et al. (2008) PLOS Genetics, showing that additive genetic variance comprises the largest component of genetic variance that contributes to phenotype, much more than gene interactions or allelic interactions “additional resources” on wiki chapter 5 is done • you should understand mutational diversity • what is a gene? a locus? an allele? • how are phenotype and genotype related? drift and selection moving into Ch 6 how fast can a population change? • 1969: French government starts spraying organophosphate insecticides along Mediterranean coast how fast can a population change? • what we know so far: populations are variable (traits, alleles)...some of this variation is heritable • not all offspring survive; those that do can pass along heritable traits • if you change the environment, the variation present may change in response Nice, France how fast can a population change? • 1969: French government starts spraying organophosphate insecticides along Mediterranean coast • mosquito population fell dramatically but started growing again by 1972 • mosquitoes near coastline were resistant to insecticide! Ester locus • encodes enzyme esterase, can detoxify organophosphates, but not normally enough to tolerate insecticide • allele called Ester esterase 1 produce more what connections can you make? • Luria-Delbruck • HIV example • straight-up natural selection at work! population genetics • you cannot follow the fate of a single allele without reference to others • the frequency of Ester 1 increased in some locations (meaning frequency of others DOWN) • we study allele frequencies through space and time, and what causes them to change evolution = change • • • • • if allele frequencies and genotype frequencies do not change, population is not evolving Hardy-Weinberg equilibrium requires a set of conditions so that population is not evolving (if population is evolving, one or more of these is NOT TRUE)... random mating with respect to locus/loci being studied population infinitely large (very big) • • • “with respect to the loci being studied” the NULL hypothesis is the boring one: that you, as a scientist, are studying a portion of the genome that is of no evolutionary interest we KNOW some traits are important for fitness and evolution, but first must reject that null hypothesis example: GENE 3000 students and 810 numbers allele frequencies • • • • • if 9 out of 10 dentists chew sugarless gum, there are 2 types of dentists, one type at frequency 0.9, the other at frequency 0.1 this is algebra: d1+d2 = 0.9 + 0.1 = 1 any symbols may be used for alleles or allele frequencies, but frequencies sum to... ONE! (1) uno un ein etc. important: may be >2 alleles genotype frequencies • genotype is just the composition of alleles describing an individual • if haploid, there is only one allele • if diploid, there are 2 • above that, it gets complicated genotype frequencies • diploid individual genotype indicated for a locus by listing both alleles, e.g. A1A1 or A1 A2 • • genotype frequency is just the proportion of individuals with that genotype, AGAIN SUMMING TO ONE (1). 1 1 AA 0.4, 1 2 A A 0.5, 2 2 A A 0.1; freqs sum to 1 • if Hardy-Weinberg holds, allele frequencies and genotype frequencies predict each other probabilities • • • • d1+d2 = 0.9 + 0.1 = 1 independent assortment of alleles: father has probability 0.9 of contributing d1, 0.1 of d2 mother same odds d1d1 genotype should be in 0.9x0.9=0.81 of offspring, d2d2 in 0.01, what about heterozygotes?