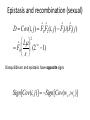* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Document
Genetic code wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Human genetic variation wikipedia , lookup
Group selection wikipedia , lookup
Species distribution wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Genetic drift wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome editing wikipedia , lookup
Genome evolution wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Oncogenomics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Microevolution wikipedia , lookup
Frameshift mutation wikipedia , lookup
Koinophilia wikipedia , lookup
Point mutation wikipedia , lookup
The Structure, Function, and Evolution of Biological Systems Instructor: Van Savage Spring 2010 Quarter 4/1/2010 Epistasis Interactions among fitness effects for different alleles Cov(wx ,wy ) w xy w x w y Can now see covariance plays central role in all of evolution. In fact, it is as central as fitness itself. If no interaction, then the covariance is 0. w xy w x w y This is know as additive (or sometimes multiplicative). Modeling more than two mutations If all mutations have the same deleterious effect, and k mutations are lethal, then ks we k ~ 1 s ~ 1 ks 1 kL k How can we modify this for epistasis? wepi 1 sk 1 ~ (1 s) k1 Lethal number of mutations sk1 ~e This measure of epistasis and definition of ε is different than in previous slide even though conceptually it is the same. Modeling more than two mutations wepi 1 sk 1 ~ (1 s) k1 sk1 ~e How do we interpret synergy and antagonism? Mutations to steps in sequence are antagonistic (green) Mutations to steps in parallel can be synthetically lethal if it knocks out a loop, which is extreme synergy (red), or multiplicative (black). Recent papers using models of epistasis: Segre, DeLuna, Church, Kishony Quantitative Epistatic Interactions Perturbation X Phenotype (Growth Rate) Perturbation Y Synergy Antagonism Suppression See also: Boone, Science (2004) Weissman, Cell (2006) Boeke, Nature Genetics (2003); Cell (2006) Giaever, Nature Genetics (2007) Measures of epistasis Since covariance is as fundamental as fitness, why not define relative covariance instead of relative fitness. We define it relative to tri-modally binned covariance that itself varies, so relative to a shifting baseline. Absolute covariance Cov(wx ,wy ) wxy wx wy Relative covariance wxy wx w y ˜ BinnedCov (wx ,w y ) w˜ xy wx w y Cov(wx ,wy ) Measures of epistasis—based onFBA predictions in yeast Sort of unimodal distribution goes to trimodal distribution Opposite of Lenki et al. because synergy is enriched. Why? Measures of epistasis—RNA viruses A bit more continuous for real data. We will see more real data later on. Higher level epistasis—interactions among functional groups rather than loci Interactions are mostly monochromatic. No reason a priori that this should be, except it signifies functional organization. Cov(WModA,WModB ) WModA,ModB WModAWModB Can we do reverse and cluster monochromatically to find functional groups? Construct network for all pairwise interactions, Start with each gene in its own group. Cluster by pairs if they interact with other genes in same way. Require monochromaticity, each group must interact with all other groups in same way Within a group there is no requirement for monochromaticity Make cluster sizes as large as possible Do clustered groups correspond to functionally annotated groups? Statistical Test: N=total gene pairs, 1034 n=interacting pairs, 278 K=same annotation pairs, 104 K N K N 1 / i n i n i 0 k1 PANN Ways to choose Ways to choose i annotated pairs n-i non-annotated pairs Ways to choose n interacting pairs Each term in sum is probability of choosing i annotated pairs out of subset of n pairs relative to any subset of n pairs being the interacting ones. Sum is probability of having k-1 or less. Can we do reverse and cluster monochromatically to find functional groups? Cluster Movie How clusterable are networks? Is clustering unique? If not, which instantiation is chosen? Number of random networks Monochromatic organization exists in the yeast metabolic network, but is very unlikely in random networks 400 Random 300 200 100 0 Yeast 0 5 10 Number of non-monochromatic connections Overlay of monochromatic and functionally annotated groups? A system-level view of modular genetic interactions in yeast metabolism P P P GLUCN GLYC P A P P P A P R R I A A I’ R R R’ A STEROL R R R’ A P P R A A I S R PENT ACAL B V M M M O M F I M URA B E F R COA ETHxt O Q’ A O Q’ IDP ATPs G H M TRPcat C J U J U PROcat J U J C L F F J U TCA L Q’ D’ J F L D L U C J J LYSbs PRObs RESPIR B T J I M C V N S B K M Q M C B T J K M C V N S B K’ M Q K C Identifying Biological Functions in Higher Hierarchical Levels GLUCN GLYC Fermentation PENT ETHxt ACAL Respiration TCA ATPs RESPIR Future directions research using digital organisms 1. I would use mixed selection criteria because I think they are more biological: a. Baseline allocation of CPU time is independent of genome size b. mathematical operations are rewarded with additional CPU time 2. Run Prism algorithm on existing digital data. Is it clusterable? What would make it clusterable? Recent papers using models of epistasis: Desai, Weissman, Feldman Mutation-selection balance μ(1-p) A1 A2 νp Given a forward mutation rate, μ, and backward mutation rate, ν pˆ ~ hs Special case that h=0, we have pˆ ~ s and Genetic Load ~ spˆ 2 Evolution of Epistasis Attempt to look at whether evolution favors additive, synergy, or antagonism Analyze evolutionary effects of different forms of epistasis in asexuals and sexuals (with recombination) with mutations Let one loci control degree of epistasis and itself be subject to selection and evolution. Not necessarily a good model because there could be mechanical, functional, and mechanistic constraints not controlled by a single or any locus. Claim there are no general patterns Assume s>>Lμ and can ignore stochastic effects Frequency distribution for number of mutations How do frequencies change in time? wF (1 L)F0 ' 0 wF wk Fk (L k)wk Fk (L 1 k)wk1Fk1 ' k (1 (L k) )e sk1 Fk (L 1 k)e Make approximations using Lμ<<s<<1 L 1 ˆ ˆ Fk ~ 1 F0 s k! k s(k1)1 Fk1 Frequency distribution for number of mutations for asexuals Normalizing our frequency distribution gives Fˆ0 ~ 1 k L L 1 s k!1 k 0 And we also have w 1 L Note there is no dependence on ε, so no selection on epistasis Additive case Fˆ0 ~ e When ε=0, notice that this is Probability of having a mutation is 1 e L s ˆ Fk ~ e k! k Frequency distribution is Poisson L s Epistasis and recombination (sexual) Proximity on genome increases recombination M is locus that controls epistasis Epistasis and recombination (sexual) D Cov(i, j) Fˆ0 Fˆ2 (i, j) Fˆ1(i) Fˆ1 ( j) L ˆ F0 (2 1) s 2 Disequilibirum and epistasis have opposite signs Sign[Cov(i, j)] Sign[Cov(w x ,w y )] Full form of disequilibrium wF (1 L)Fˆ0 r( j i)D(i, j) ' 0 i j Same basic form as we already derived in class 2 Lr 1 j i L 2i 2 j 2 2 2 ˆ (i, j) (2 1)Fˆ 1 D Fˆ0 (2 ( 1) ( 1) 0 s s L L s 2 2 Recombination decreases disequilibrium but does not change its sign. Recombination-selection balance and mutation-selection balance come in similar forms Distribution of mutations Synergistic population favors recombination because it breaks up mutations to increase fitness. Opposite for antagonistic population Time evolution of fitness for mixed (synergy and antagonism) population L Lr 1 2 w w1 w 0 ~ R 1 1 s 3 2 2 R is recombination between epistasis locus and rest of loci. Population 1 is antagonistic and population 0 is synergistic. Comparison of analytical and simulations Does recombination favor synergy or antagonism? Average fitness is same. Antagonistic individuals will build up mutations and recombination will shift those to synergistic individuals and dramatically decrease their fitness. Starting with a synergistic population, recombination would be able to take off and establish. Once established, it would push the population towards more antagonistic epistasis, which might then help eliminate recombination.