* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Maternal effect genes
Oncogenomics wikipedia , lookup
RNA interference wikipedia , lookup
Essential gene wikipedia , lookup
Point mutation wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epitranscriptome wikipedia , lookup
Messenger RNA wikipedia , lookup
Primary transcript wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Designer baby wikipedia , lookup
Minimal genome wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epigenetics of human development wikipedia , lookup
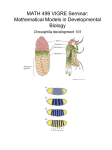
Animal Development Drosophila axis formation Part 1: A-P patterning [email protected] Problem: starting point where all cells have the same developmental potential (because they have the same DNA) However, the end point is the production of nerve cells, muscle cells, epithelial cells etc. Therefore differentiation happens. Mechanism? First Step: Breakage of symmetry Inside Cells Diagrams representing a group of mesenchymal cells that emit a signal (black dots) Salazar-Ciudad I Development 2010;137:531-539 In this lecture: The origin of Anterior-Posterior Axis Mutant screens to isolate segmentation genes Genetic analysis of early acting determinants Important roles of post-transcriptional regulation and mRNA/protein localisation Methods of dissecting enhancers Dosage-dependent activation of zygotic genes Hierarchical organisation of segmentation genes THEY LIVE…. Developmental biology: Drosophila segmentation and repeated units * egg: generate the system * larva: eat and grow * pupa: structures in larvae grow out to form adult fly: metamorphosis (Drosophila is a holometabolous insect) 1 The chromosomal basis of inheritance Mapping genes onto chromosomes via recombination Wing mutant A Eye mutant C Leg mutant B 20 mu 40 mu The genetic map The early embryo is a syncitium The unusual feature of the Drosophila early embryo is that the first 13 mitoses are nuclear divisions without concomitant cytoplasmic division, making the embryo a syncitium-a multinucleated cell. After division 9, the plasma membrane of the oocyte evaginates at the posterior pole to surround each nucleus thus creating the pole cells, which will form the fly’s germ line. Segments in embryos are maintained throughout development Forming complex pattern: establishing positional information The Hunt for Mutants 30,000 independently-derived mutants in genes required for survival. 8,000 mutants define genes required for embryonic survival (these became the focus the study). 750 mutants have specific effects on A/P or D/V patterning. 150 genes with specific effects on A/P or D/V patterning identified by the 750 mutants (average of ~ 5 alleles per gene). A Detour Into Embryonic Anatomy – Denticle Bands Denticle bands on a 1st instar larva. Denticle bands are hair-like projections on the ventral cuticle of an embryo. Denticle bands provide an easily visualized marker of embryonic/larval pattern. Maternal effect genes • Phenotype of the embryo is determined by the genotype of the mother. • The polarity and spatial coordinates of the embryo are initially set by the products of these genes (therefore, sometimes called “coordinate genes”). • The gene products, either mRNA transcripts, proteins, or cell surface ligands are contributed by the nurse cells or follicle cells as the egg is constructed. • The dorsal-ventral axis (1 gene-system, 12 genes) and anteriorposterior axis (3 gene-systems; anterior, 4 genes, posterior, 11 genes, and terminal, 6 genes) determined by maternal effect genes. • Originally isolated as homozygous mutant, adult females that lay normal looking eggs that do not develop at all, regardless of the genetic contribution of the male. Four Independent Genetic Regulatory Systems Specify the Anteroposterior and Dorsoventral Axes Maternal effect genes • All four systems share several properties: (i) the product of (at least) one gene is localized in a specific region of the egg, (ii) this spatial information results (directly or indirectly) in an asymmetrical distribution of a transcription factor, (iii) the transcription factor is distributed in a concentration gradient that defines the limits of expression of one or more zygotic target genes, such as segmentation genes. Bicoid mutant embryos lack head and thorax structures +/+ mother bicoid/bicoid mother The Importance of RNA localisation Essential for many fundamental processes: • Cell polarity • Developmental patterning • Neural development • Learning and memory Hamilton et al 2012, Biophysics for the life sciences Chapter 11 Establishment of AP axis in oogenesis and bicoid localisation by Gurken signalling. In early stage egg chambers MTOC is in the oocyte, and gurken mRNA is localised at posterior. Translation and limited diffusion means signal sent to overlying posterior follicle cells (received via torpedo receptor). A signal is sent back which activates protein kinase A in the egg oocyte cytoskeleton is re-organised and directs the localisation of bicoid and oskar, defining the A-P axis. EGF signalling between the oocyte nucleus and follicle cells Isolating the ovary Ovary Weil, Parton, Davis, Jove (2012) Stage 9 Egg Chamber nurse cell follicle cells oocyte grk mRNA bcd mRNA osk mRNA Weil, Parton, Davis, Jove (2012) Effect of replacement of the 3’ UTR of the nos mRNA with the 3’ UTR of bcd mRNA • The nos-bcd transgene is able to localise at the anterior pole and as a consequence NOS protein will inhibit translation of the hb and bcd mRNAs. The Bcd gradient mRNA (in situ) Protein (Ab staining) Concentration gradients of BCD and HB-M establish A-P axis • Positional information along the A-P axis of the syncitial embryo is initially established through the creation of concentration gradients of two transcription factors: Bicoid (BCD) and Hunchback (HB-M). These are products of two maternal effect genes their mRNAs provided by the mother and stored in the embryo until translation initiates. These factors interact to generate different patterns of gene expression along the axis. Bicoid is an Anterior Morphogen Note that bicoid (and other maternal effect gene products) diffuse in the shared cytoplasm of the syncytial blastoderm. This is a unique feature of insect embryogenesis. BCD acts as a concentration-dependent manner Thresholds can turn gradients into sharp boundaries – Bicoid protein required for early activation of zygotic hunchback. – Bicoid contains homeobox – Mutations in homeobox results in failure of Bicoid protein to interact with hunchback target sequences. bicoid protein gradient – gradient is interpreted at least at four different levels (thresholds). bicoid as a repressor of posterior fates Bicoid binds the 3’ UTR of caudal mRNA and suppresses translation. Caudal protein Caudal protein enters the nuclei at the posterior end of the syncytial blastoderm and helps specify posterior fates At the Posterior: nanos localisation by Gurken signalling oocyte cytoskeleton is reorganised and directs the localisation of bicoid and oskar, defining the A-P axis. oskar mRNA binds Kinesin I and Staufen proteins. Kinesin I localises oskar mRNA to posterior Staufen allows translation of oskar mRNA Oskar protein binds nanos Effect of posterior group genes on hunchback. Posterior group genes: • primary gene is nanos. • nanos mRNA is tightly localised to the posterior pole of the egg. Effect of posterior group genes on hunchback. The role of nanos is to disable hb maternal mRNA at the posterior end of the egg. Four Independent Genetic Regulatory Systems Specify the Anteroposterior and Dorsoventral Axes Terminal group genes Torso: TRK signalling via MAPK Segmentation pattern Obvious segmentation begins to develop by germ band extension stage. The embryonic segmentation pattern has direct analogs to the final segments of the adult. Segmentation pattern can be thought of as classical segments or midsegment-tomidsegment intervals called parasegments. Some early embryonic segments become incorporated into the complex structures of the head and mouth. SUMMARY Nurse cells surrounding the oocyte in the ovarian follicle provide it with large amounts of mRNAs and proteins, some of which become localised in particular sites. The oocyte produces a local signal, which induces follicle cells at one end to become posterior follicle cells. The posterior follicle cells cause a re-organisation of the oocyte cytoskeleton that localises bicoid and hunchback mRNA to the anterior end and other mRNAs such as oskar and nanos to the posterior end of the oocyte. Following fertilisation, development starts and these mRNAs are translated. Subsequently, gradients of the BCD and HB proteins define the anterior nuclei-the embryo is still a syncytial blastoderm, while inhibition of translation of their mRNAs by Nanos define the posterior cells. Nuclei in between receive a variable amount of BCD and HB resulting in differential activation or repression of target genes and finally in different developmental cell fates.