* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 幻灯片 1 - Wiley
Gene nomenclature wikipedia , lookup
Genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Population genetics wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Koinophilia wikipedia , lookup
History of genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Frameshift mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene expression programming wikipedia , lookup
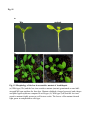
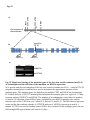
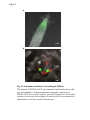
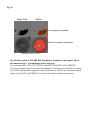
Fig. S1 a Ws mutant b Fig. S1 Morphology of the low-iron-sensitive mutant of Arabidopsis. (a) Wild type (Ws) and the low-iron-sensitive mutant (mutant) germinated on one-halfstrength MS agar medium for four days. Mutant exhibited a longer hypocotyl and a larger and paler hypocotyldeons compared to wild type. (b) Wild type (left) and the low-ironsensitive mutant (right) grown on soil for two weeks. The leaves of the mutant showed light green in comparison to wild type. Fig. S2 NGA8 Chromosome 4 Recombinant SSLP3624 SSLP1475 CIW5 a c 5 1 0 3 med16-4 GT-AT b ATG TAG med16-2 med16-3 c Ws med16-4 AtMED16 AtACTIN8 Fig. S2 Map-based cloning of the mutation gene of the low-iron sensitive mutant (med16-4) of Arabidopsis and the affection of the mutation on MED16 expression. (a) A genetic and physical mapping of the low-iron sensitive mutant (med16-4) . A total of 326 F2 progenies homozygous for mutant were used to determine the approximate position of the mutation gene. The mutation gene was linked to the markers CIW5 and NGA8 on chromosome IV. Further analysis of 2876 F2 mutant plants delimited the mutation gene to a region of 3.1 Mbp between markers SSLP1475 and NGA8. The number of recombination events linked to markers is marked. (b) Structure of the MED16 gene (At4g04920), the MED16 mutation site, and the insertion sites of the T-DNA in sfr6-2 (med16-2) and sfr6-3 (med16-3). The filled boxes represent exons and the lines indicate introns. (c) RT-PCR analysis of MED16 expression in med16-4 mutant. ACTIN8 was used as loading control. RNAs were extracted from seedlings grown on onehalf-strength MS agar medium with iron for 10 days. Fig. S3 a root b leaf Fig. S3 Subcellular localization of Arabidopsis MED16. The plasmid 35S:MED16-GFP was constructed and transferred to wild type of Arabidopsis. Transformed plants with stable expression of MED16-GFP were selected, and the green GFP signal were observed in root tips of one-week-old seedlings (a) and in leaf (b) of one-month-old plants under a Carl Zess confocol microscope. Fig. S4 WT sin4 MED16 ACT1 Fig. S4 Characterization of the yeast MED16/SIN4-knocking out mutant. RT-PCR analysis of knocking out of SIN4 (a MED16 homolog in yeast genome) in sin4 yeast mutant, ACT1 was used as loading control. Fig. S5 Bright Field Epifluo Col mesophyll protoplasts a med16-3 mesophyll protoplasts b bHLH38-CC/FIT-YN Fig. S5 Observation of FIT/bHLH38 heterodimer formation in mesophyll cells of the mutant med16-3 of Arabidopsis and its wild type. The constructs BiFC-FIT-nYFP (FIT-YN) and BiFC-AtbHLH38-cCFP (bHLH38CC)were introduced into the mesophyll protoplasts of wild type (a) and med16-3 mutant (b) by PEG transformation approach. After incubation for 16-20 h, the interaction signal (green color) of FIT with bHLH38 were observed under a confocol microscope. Fig. S6 a WT med16-3 med16-3ox29/38 MED16 ACTIN8 b ATG TAG TGG-TAG c WT med16-3 med16-5ox29/38 MED16 ACTIN8 med16-5ox29/38 Fig. S6 The med16 mutants (med16-3ox29/38 and med16-5ox29/38) of Arabidopsis in the genetic background of ox29/38. (a) RT-PCR verification of lacking MED16 expression in med16-3ox29/38 mutant generated by crossing med16-3 with ox29/38, ACTIN8 was used as loading control. (b) Gene structure diagram of MED16 and the position of the point mutation from TGG to TAG in the mutant line med16-5ox29/38 obtained by screening a EMS mutation population of ox29/38. (c) RT-PCR analysis of MED16 expression in med16-5ox29/38, ACTIN8 was used as loading control.