* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download PDF
Epigenetics of human development wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Preimplantation genetic diagnosis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Designer baby wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
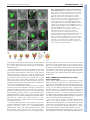
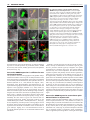
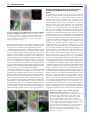
RESEARCH ARTICLE 117 Development 138, 117-126 (2011) doi:10.1242/dev.059147 © 2011. Published by The Company of Biologists Ltd Coordination of apical and basal embryo development revealed by tissue-specific GNOM functions Hanno Wolters*, Nadine Anders†, Niko Geldner‡, Richard Gavidia and Gerd Jürgens§ SUMMARY Flowering-plant embryogenesis generates the basic body organization, including the apical and basal stem cell niches, i.e. shoot and root meristems, the major tissue layers and the cotyledon(s). gnom mutant embryos fail to initiate the root meristem at the early-globular stage and the cotyledon primordia at the late globular/transition stage. Tissue-specific GNOM expression in the gnom mutant embryo revealed that both apical and basal embryo organization depend on GNOM provascular expression and a functioning apical-basal auxin flux: GNOM provascular expression in gnom mutant background resulted in non-cell-autonomous reconstitution of apical and basal tissues which could be linked to changes in auxin responses in those tissues, stressing the importance of apical-basal auxin flow for overall embryo organization. Although reconstitution of apical-basal auxin flux in gnom results in the formation of single cotyledons (monocots), only additional GNOM epidermal expression is able to induce wild-type apical patterning. We conclude that provascular expression of GNOM is vital for both apical and basal tissue organization, and that epidermal GNOM expression is required for radial-to-bilateral symmetry transition of the embryo. We propose GNOM-dependent auxin sinks as a means to generate auxin gradients across tissues. INTRODUCTION The basic body organization of the plant seedling is laid down during embryogenesis, which, in Arabidopsis, is characterized by highly reproducible cell division patterns (Fig. 1Q) (Jürgens and Mayer, 1994). The apical-basal organization of the embryo is initiated after the asymmetric division of the zygote. The large basal cell will form the extra-embryonic suspensor and part of the root meristem, the apical cell will give rise to the pro-embryo. At the octant stage, periclinal divisions of the pro-embryo initiate the radial organization of the embryo, generating an outer protodermal layer and an inner cell mass. At the early-globular stage, divisions of these inner cells produce an outer ground-tissue layer and a centrally located provascular tissue. Root-meristem formation is initiated shortly afterwards by the asymmetric division of the hypophysis located at the basal pole of the embryo. At the lateglobular/transition stage, the cotyledon primordia are initiated, changing the symmetry of the embryo from radial to bilateral (Fig. 1Q). Most patterning processes are completed by the late globular/transition stage and subsequent stages are characterized primarily by massive growth of the embryo. Many gnom mutant alleles were isolated in a screen for seedlings with disturbed organ formation (Jürgens et al., 1991). gnom mutants lack a primary root, the cotyledons are fused or completely missing, and the mutant seedlings remain small, showing neither hypocotyl nor cotyledon elongation. During ZMBP, Entwicklungsgenetik, Universität Tübingen, Auf der Morgenstelle 3, 72076 Tübingen, Germany *Present address: Developmental Biology, EMBL Heidelberg, Meyerhofstr.1, 69117 Heidelberg, Germany † Present address: University of Cambridge, Department of Biochemistry, Tennis Court Road, Cambridge CB2 1QW, UK ‡ Present address: Plant Cell Biology Laboratory, DBMV, Biophore, UNIL-Sorge, University of Lausanne, 1015 Lausanne, Switzerland § Author for correspondence ([email protected]) Accepted 27 October 2010 embryogenesis, gnom mutant defects appear as early as at the division of the zygote (Mayer et al., 1993). At the globular stage, the root pole is not correctly formed and inner tissue organization is strongly perturbed. The longitudinal cell files observed in the wild-type embryo are disrupted and the orientation of cell divisions is rather random. Additionally, the transition to the bilateral symmetry does not take place giving the gnom heart-stage embryo a ball-shaped appearance (Mayer et al., 1993). The GNOM gene encodes an ARF guanine-nucleotide exchange factor (ARF-GEF) required for vesicle budding from donor membranes (Busch et al., 1996; Steinmann et al., 1999). ARF-GEFs activate ARF (adenosine ribosylation factor) proteins by exchanging ARF-bound GDP for GTP and thereby initiate recruitment of vesicle coat proteins (Casanova, 2007). GNOM, together with its closest homologue GNOM-LIKE 1 (GNL1), is required for ER-Golgi trafficking, but in contrast to GNL1, GNOM has an additional plant-specific role in endosomal recycling of auxin-efflux carrier PIN1 to the basal plasma membrane (Geldner et al., 2003; Richter et al., 2007). This recycling is necessary for the coordinated polar localization of PIN1 and therefore for polar auxin transport (Steinmann et al., 1999; Geldner et al., 2003). The genetic elimination of embryonically expressed auxin-efflux carriers PIN1, PIN3, PIN4 and PIN7, or treatment of wild-type embryos with auxin transport inhibitors, indeed mimic the gnom mutant phenotype (Friml et al., 2003; Hadfi et al., 1998). Here, we analyze the role of GNOM in apical-basal and radial patterning of the embryo. Using a transactivation approach, we restrict GNOM expression to specific embryonic domains of gnom mutant embryos, thus generating genetic mosaics, and analyze the effect on overall embryo patterning. We show that limited GNOM expression has direct effects on the tissue of expression. Surprisingly, we also observe reestablishment of normal patterning in GNOM non-expressing tissues. We also provide evidence that the remote control of apical and basal embryo patterning by GNOM is directly linked to apical-basal auxin flow because all observed non-cell-autonomous GNOM effects are preceded by a DEVELOPMENT KEY WORDS: Embryo patterning, Auxin, GNOM, Arabidopsis RESEARCH ARTICLE non-cell-autonomous reconstitution of the PIN1 carrier. We propose a patterning model in which GNOM expression induces an ‘auxin sink’, depleting auxin from GNOM non-expressing tissues. We propose that GNOM-dependent PIN relocalization (cell autonomous) and sink-driven auxin transport (non cellautonomous) trigger the initiation of auxin transport routes in embryonic pattern formation. MATERIALS AND METHODS Plasmid construction GNOM reporter plasmid for the LhG4>>pOp two-component expression system This has previously been described by Moore et al. (Moore et al., 1998). The c96 vector carrying the GNOM-3xmyc cDNA (Geldner et al., 2003) was SalI digested and the GNOM-3xmyc cDNA inserted into the XhoI site of ML208 (obtained from M. Lenhard, University of Potsdam, Germany), yielding ML208 pOp::GNOM-3xmyc, carrying the GNOM cDNA behind a 2xOp sequence. The ML208 vector carries an additional 2xOp::GUS reporter to monitor GNOM reporter gene expression. GNOM reporter plasmid for the Gal4:VP16 >> UAS twocomponent expression system This has previously been described by Haseloff and Laplaze et al. (Haseloff, 1999; Laplaze et al., 2005). The SalI fragment of c96 GNOM3xmyc was inserted into the SalI site of pGRIIB UAS tNOS (Weijers et al., 2006), yielding pGRIIB UAS::GNOM-3xmyc. The resultant vectors were transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998) and T1 transformants were selected with BASTA (Bayer AG, Germany). Plant material The homozygous LacI:Gal4 (LhG4) driver lines AtHB8 and ANT were kindly provided by M. Lenhard (Alvarez et al., 2006; Schoof et al., 2000), the RPS5a driver line by R. Groß-Hardt (ZMBP, University of Tübingen, Germany), the LTP driver line by P. Gallois (University of Manchester, Manchester, UK) (Baroux et al., 2001) and the pOp::GFP reporter by Y. Eshed (Weizmann Institute of Science, Rehovot, Israel) (Lifschitz et al., 2006). The homozygous GAL4:VP16 driver lines N9217, N9094, N9135, N9249 and N9312 were generated by Jim Haseloff (University of Cambridge, Cambridge, UK) and obtained from the Nottingham Arabidopsis Stock Centre (http://www.plantsci.cam.ac.uk/Haseloff/) (Haseloff, 1999; Laplaze et al., 2005; Scholl et al., 2000). The ANT, LTP, N9217 driver and pOp::GFP lines have been previously described (Alvarez et al., 2006; Schoof et al., 2000; Baroux et al., 2001; Weijers et al., 2006; Lifschitz et al., 2006). The SHR::SHR-GFP marker was kindly provided by P. Benfey (Duke University, Durham, NC, USA) (Nakajima et al., 2001), TCS::GFP by B. Müller (Institute of Plant Biology, University of Zürich, Switzerland) (Müller and Sheen, 2008), SCR::GFP by A. Schlereth (Syngenta Forschungszentrum, Stein, Switzerland), DR5rev::GFP by J. Friml (VIB, Gent, Belgium) (Friml et al., 2003), DR5rev::3xVenus-N7 by S. Gordon (California Institute of Technology, Pasadena, CA, USA) (Heisler et al., 2005) and PIN4 antibody by K. Palme (University of Freiburg, Freiburg, Germany) (Friml et al., 2002). The strong gnom alleles emb30-1 and xg33 named ‘gnom’ in the following were used as the mutant background for both marker analysis and transactivation of GNOM (Shevell et al., 1994; Busch et al., 1996). Because the marker/promotor expression pattern in gnom did not always match with the expression pattern in wild type, each of the transactivator lines was carefully analyzed in gnom before the rescue analysis was performed. Growth conditions and generation of transactivation lines Plants were grown in a 16 hour light/8 hour dark cycle at 18°C or 23°C. For better distinction of the seedling rescue phenotypes seeds were generally grown on MS plates for 2-3 weeks before analysis. Driver (Promoter) and reporter (GNOM) lines were first crossed into the gnom mutant background (xg33 or emb30-1). gnom heterozygous driver and reporter lines were subsequently crossed to one another. The resultant Development 138 (1) F2 and F3 transactivation lines were grown on selective plates for test of homozygosity of both driver and reporter insertions and heterozygosity of gnom. Transactivation was checked by GUS staining and GNOM expression in western blots (with anti-myc monoclonal antibody). F3 plants homozygous for both driver and reporter constructs (Promoter/Promoter GNOM/GNOM gnom/+) yielded 100% GUS-positive seedlings. Antibody staining and confocal laser-scanning microscopy A pOp::GFP reporter (obtained from Y. Eshed) (Lifschitz et al., 2006) was crossed into all LhG4>>pOp transactivation lines to monitor spatiotemporal GNOM expression. F2 and homozygous F3 lines carrying the GFP reporter (in the case of LhG4>>pOp system) were analyzed by confocal laser scanning microscopy [Leica TCS SP2 (Wetzlar, Germany)]. Images were processed using LAS AF Lite (Leica Microsystems) or Adobe Photoshop CS2. Where mentioned, transactivation lines were crossed with the DR5rev::GFP reporter (Friml et al., 2003) or DR5rev::3xVenus-N7 (Heisler et al., 2005). In several cases (ANT, AtHB8, LTP), GFP reporter expression was also analyzed by GFP antibody staining to increase the detection level of the GFP reporter in early embryos. Immunofluorescence preparations were carried out as described previously (Lauber et al., 1997). Antibodies and dilutions used were as follows: c-myc 9E10 monoclonal antibody (1:600; Santa Cruz); rabbit anti-PIN1 (1:800) (Friml et al., 2003); rabbit anti-PIN4 (1:400) (Friml et al., 2002); monoclonal mouse anti-GFP (1:600; Roche); rabbit anti-GFP (1:600; Molecular Probes); CY3 antirabbit (1:600; Dianova); Alexa488 anti-mouse (1:600; Invitrogen) and Alexa488 anti-rabbit (1:600; Invitrogen). RESULTS Marker analysis in gnom mutant embryos The mutant phenotype of gnom was first described by Jürgens et al. (Jürgens et al., 1991). gnom mutant embryos fail to properly form a root and shoot apical meristem, and bilateral symmetry is not established, resulting in a ball-shaped embryo. To better understand the origin of the patterning defects in gnom, we sought to identify early events of tissue misspecification by introducing tissue-specific markers into gnom and comparing the resulting expression patterns with those of wild-type control embryos. Both SHORTROOT (SHR) and SCARECROW (SCR) are required to set up the post-embryonic root meristem niche (Di Laurenzio et al., 1996; Sabatini et al., 2003). Radial defects in the shr and scr mutant can be traced back to the embryo, indicating that SHR and SCR specify endodermal fate during embryogenesis (Scheres et al., 1995; Wysocka-Diller et al., 2000; Sabatini et al., 2003). Because the root meristem is not established in gnom, we wondered whether SHR expression is also altered. Interestingly, although in early gnom embryos provascular SHR::SHR-GFP expression (Nakajima et al., 2001) was still detected, SHR::SHRGFP expression in heart-stage gnom embryos was restricted to the apical/central domain (Fig. 1A,E). Importantly, SHR::SHR-GFP expression was excluded from the root meristem region. However, SHR movement to the adjacent cell layer was not impaired in gnom. Similarly, SCR::GFP expression, which labels the hypophysis and the ground tissue precursors in wild-type embryos (Wysocka-Diller et al., 2000) (Fig. 1B), was lost or very weak at the basal pole of gnom embryos. However, scattered SCR expression in the ground tissue precursors above the root pole often remained (Fig. 1F). In rare cases (2/44), SCR expression was confined to the apical end, which was never observed in wild-type embryos (data not shown). In summary, although in globular gnom embryos both vascular SHR::SHR-GFP as well as weak SCR::GFP signals in the root meristem region have been observed, SHR and SCR expression were lost from the root pole region by the heart stage. Another marker for the embryonic root meristem niche, the cytokinin-responsive TCS::GFP (Müller and Sheen, 2008), DEVELOPMENT 118 Auxin transport and embryo patterning RESEARCH ARTICLE 119 Fig. 1. Changes in marker expression observed in the gnom mutant embryo. Confocal analysis of the expression pattern of diverse driver lines (N9094, N9135, N9217, ANT, AtHB8) and tissue-specific markers (SHR, SCR, TCS) in wild type (A-D,I-L) and in gnom mutant embryos (E-H,M-P). (A,E)SHR::SHR-GFP (heart stage); (B,F) SCR::GFP (heart stage). SHR and SCR are absent or only weakly expressed in the root meristem region in gnom. (C,G)TCS::GFP (late globular (G) and heart (C) stage). (D,H)ANT>>GFP (heart stage), (I,M) N9094>>GFP (heart stage). (J,N)N9135>>GFP (globular (J) and early heart (N) stage). (K,O)N9217>>GFP (globular stage). (L,P)AtHB8>>GFP (late globular stage). Scale bar: 10m. (Q,R)Schematic overview of embryo development and expression domains of selected markers and driver (promoter) lines. (Q)Stages of wild-type embryogenesis (from left to right): dermatogen, earlyglobular, mid-globular, late-globular/transition and mid-heart. (R)Transition/early heart stage of gnom development. Expression domains are highlighted in color: provasculature, AtHB8, N9217 (pink); hypophysis and its derivatives, N9312 (light blue); cortex/endodermis cells, N9094, N9135, SCR (red); cotyledon founder cells, ANT (green); protoderm, LTP (yellow); suspensor, N9249 (dark blue). and N9094 expression was observed in mp and N9094 radial expression was also maintained in mp nph4 mutants (see Fig. S2D-I in the supplementary material). We cannot exclude the possibility that other ARFs still functioning in the embryo might be required for radial embryo patterning, but the intact radial organization in gnom and mp nph4 suggests that apical-basal auxin transport is not required to maintain the radial organization of the embryo. RPS5a>>GNOM fully complements the gnom mutant phenotype Correct embryo patterning involves communication between different cells and tissues (Weijers et al., 2006; Breuninger et al., 2008). To dissect and identify the origin of the defects in the gnom mutant embryo, we decided to express GNOM in defined domains in the gnom null mutant background using a transactivation approach (Moore et al., 1998; Haseloff, 1999). (In the following sections, GNOM transactivation from different promoters is symbolized by ‘promoter >> GNOM’.) This approach allowed us to identify the role of GNOM in specific embryonic tissues, as well as to analyse possible GNOM-dependent signaling to neighboring tissues (‘non-cell-autonomous’ GNOM function). To test our system, we used the ribosomal RPS5a promoter. Similar to the GNOM promoter (see Fig. S1A-D in the supplementary material) (Geldner et al., 2004) the RPS5a promoter is expressed in all tissues during early embryogenesis (see Fig. S1E-F in the supplementary material) (Weijers et al., 2001). GNOM transactivation from the RPS5a promoter (RPS5a >> GNOM) fully rescued the gnom mutant phenotype (see Fig. S1G-I in the supplementary material), demonstrating the functionality of DEVELOPMENT showed only weak expression at the basal pole of gnom, whereas the suspensor signal also seen in wild type is maintained (Fig. 1C,G). These data indicate that the root pole is not properly established in gnom. As cotyledon formation in gnom is also impaired, we next tested whether apical markers were correctly expressed in gnom. The apical marker AINTEGUMENTA>>GFP (ANT>>GFP), which in wild-type heart-stage embryos is expressed throughout the cotyledon primordia (Long and Barton, 1998; Schoof et al., 2000), was detected around the whole apical circumference of gnom embryos (Fig. 1D,H), and was mainly restricted to the epidermis. This aberrant marker expression reflects the failure of gnom to establish a bilateral symmetry that results in fused cotyledons (see Fig. S1H in the supplementary material). By contrast, the radial markers N9094 and N9135, which label the embryo ground tissue/endodermis, and N9217 (also known as Q0990) and AtHB8, which label the provasculature, are expressed normally in gnom embryos (Fig. 1I-P). This is in line with earlier results showing wild-type expression of the epidermal marker MERISTEM LAYER1::3xGFP in gnom (AtML1::3xGFP) (Takada and Jürgens, 2007). In summary, although apical and basal markers either did not maintain their initial expression or were misexpressed, expression of radial markers was not affected in the gnom mutant (Fig. 1R). Given that radial markers are correctly expressed in gnom, we wondered whether this would also be the case for other mutants affecting embryonic auxin flow. NONPHOTOTROPIC HYPOCOTYL4 (NPH4) and MONOPTEROS (MP) are ARF auxin response factors and both have been implicated in embryonic patterning (Hardtke et al., 2004). Wild-type N9217 120 RESEARCH ARTICLE Development 138 (1) Fig. 2. Rescue analysis of gnom mutants expressing GNOM from a provascular promoter. Confocal and light microscopic analysis of N9217>>GNOM and AtHB8>>GNOM transactivation lines. (A)N9217>>GFP in gnom (early heart stage). (B)N9217>>GFP in N9217>>GNOM gnom (heart stage). (C,D)Segregating N9217>>GNOM gnom seedlings from lines expressing one (C) or two (D) copies of N9217>>GNOM (representative phenotypes selected: see Table S1 in the supplementary material). (E)AtHB8>>GFP in gnom (late globular stage). (F)AtHB8>>GFP in AtHB8>>GNOM gnom embryo (heart stage). (G,H)Segregating AtHB8>>GNOM gnom seedlings from lines expressing one (G) or two (H) copies of AtHB8>>GNOM (see also Table S1 in the supplementary material). (I,J)PIN1 localization (red) in wild-type (I) and gnom (J) heart stage embryo. (K,L)DR5rev::GFP expression in late-globular wild-type embryo (K) and in globular gnom embryo (L). (M-R)PIN1 localization (red) in N9217>>GNOM (M-P) and AtHB8>>GNOM (Q,R) gnom mutant embryos also carrying a GFP reporter. (N,P,R) High-magnification views of marked regions in M,O,Q, respectively. Basal PIN1 localization is observed above the GNOM expression zone. (S,T)DR5rev::GFP in segregating N9217>>GNOM gnom globular-stage embryos from lines expressing one (S) or two (T) copies of N9217>>GNOM. The DR5 signal at the embryo apex is strongly reduced in N9217>>GNOM homozygous lines. Scale bar: 1 mm. Provascular GNOM expression is sufficient for root meristem formation The asymmetric division of the hypophysis in the globular embryo initiates the formation of the root pole. In embryos defective in the auxin response factor, MONOPTEROS, or the auxin (IAA)-induced Aux/IAA family member BODENLOS, this asymmetric division is disturbed (Hamann et al., 1999; Berleth and Jürgens, 1993). Interestingly, both MONOPTEROS and BODENLOS are not expressed in the hypophysis itself but rather in the provascular cells adjacent to the hypophysis (Hamann et al., 2002). When we expressed GNOM exclusively in the provasculature, above the future root meristem region, from the dermatogen stage onwards by using the driver line N9217 (also known as Q0990) (Weijers et al., 2006), N9217>>GNOM gnom embryos showed a normal basal meristem region and later on developed a primary root (Fig. 2A-D). Essentially the same rescue of gnom was observed when we expressed GNOM from another provascular promoter of the HOMEOBOX8 gene (AtHB8>>GNOM) (Baima et al., 1995) (Fig. 2E-H). Importantly, GNOM expression in the hypophysis and its derivatives, which will be part of the future root meristem, was not required for the functional reconstitution of the root meristem in gnom embryos, arguing for a non-cell-autonomous action of provascular GNOM expression on root meristem formation and/or maintenance. Strikingly, GNOM expression from the provascular promoter, N9217, not only reconstituted the root meristem region of gnom but also promoted normal hypocotyl elongation and partial reconstitution of cotyledons (Fig. 2C,D). The rescued seedlings often had only one or fused cotyledons, but otherwise showed cotyledon expansion and partially rescued vascular organization, which was never observed in gnom seedlings (see Fig. S3C,D in the supplementary material). An even stronger reconstitution of apical tissues was mediated by GNOM expression from the AtHB8 provascular promoter (Fig. 2H). However, the AtHB8 promoter is also active in the apical epidermis, unlike the provascular promoter N9217 (Fig. 1L,P; see Discussion). In summary, GNOM expression exclusively in inner provascular cells of gnom is sufficient to induce embryonic root meristem formation as well as apical-basal embryo elongation, and partial reconstitution and elongation of cotyledons. Both the basal and the apical rescue effects seem to be triggered non-cell-autonomously by GNOM expression in a central region. Provascular GNOM expression reconstitutes polar PIN localization in adjacent cells GNOM is required for PIN-FORMED1 (PIN1) recycling and for auxin transport (Steinmann et al., 1999; Geldner et al., 2003). In wild-type embryos, the PIN1 auxin-efflux carrier is localized to the basal membrane of provascular cells (Steinmann et al., 1999) (Fig. 2I). By contrast, PIN1 is not polarly localized in gnom embryos (Steinmann et al., 1999) (Fig. 2J), and auxin transport to the basal pole is disrupted, leading to presumptive auxin accumulation, which is supported by the strong DR5rev::GFP signal in the apical DEVELOPMENT the transactivation system. In the following, we analyze patterning responses to GNOM expression in provascular (N9217, AtHB8 driver lines), basal (N9312, N9249), apical (ANT) and epidermal (LTP) tissues (Fig. 1Q). Fig. 3. Rescue analysis of gnom mutants expressing GNOM from a basal promoter. Confocal and light microscopic analysis of N9249>>GNOM and N9312>>GNOM transactivation lines. (A,B)N9249>>GFP in globular stage wild-type (A) and gnom (B) embryo; (C) N9249>>GNOM gnom seedling; (D,E) N9312>>GFP in globular stage wild-type (D) and heart stage gnom (E) embryo; (F) N9312>>GNOM gnom seedling; (G,H) PIN1 immunofluorescence staining (red) in N9312>>GNOM gnom embryos (heart stage) also carrying a GFP reporter. (H)high-magnification view of marked region in G. Strong polar PIN1 localization above the GNOM expression zone. (I)DR5::3xVenus-N7 expression (green) in N9312>>GNOM gnom embryo (heart stage). Scale bar: 0.2 mm in E; 1 mm in F. region of globular-stage gnom embryos (Friml et al., 2003) (Fig. 2K,L). When we expressed GNOM from the provascular promoters N9217 or AtHB8, we observed a reconstitution of PIN1 polar localization in the provasculature and also parts of the apical region of gnom mutant embryos (Fig. 2M-R). Concomitantly, apical auxin accumulation, reflected by the DR5rev::GFP reporter activity (Friml et al., 2003), was strongly reduced (compare Fig. 2L with 2S,T). Subsequently, the N9217>>GNOM and AtHB8>>GNOM rescued gnom mutants were analyzed in more detail to reveal the PIN1 localization characteristics at GNOM expression boundaries. Surprisingly, we found a clear basal localization of PIN1 in nonGNOM expressing apical cells directly adjacent to the GNOM expression domain (Fig. 2N,P,R). This suggests that GNOM expression in lower cells is sufficient to induce correct PIN1 localization and thereby a strong directional auxin flow in directly adjacent upper tissues. Hypophyseal GNOM expression reconstitutes the apical-basal embryo axis The non-cell-autonomous rescue effects in N9217>>GNOM rescue mutants prompted us to analyse the impact of GNOM expression exclusively in the basal region of the embryo. To do this, we made use of the N9249 and N9312 promoter lines, which confer expression in the suspensor or the hypophysis region, respectively (Fig. 3A,B,D,E). During late stages of embryogenesis (torpedo stage) N9312>>GFP expression was also seen in the shoot meristem region and weakly in the vasculature (not shown). If GNOM function were restricted to the cells where it is expressed, basal GNOM expression would have no effect on gnom central and RESEARCH ARTICLE 121 apical defects. Strikingly, whereas GNOM expression in the suspensor did not result in a discernible rescue of the gnom mutant phenotype (Fig. 3C), GNOM expression (additionally) in the hypophysis resulted in a significant portion of gnom seedlings with a rescued primary root and elongated hypocotyl (Fig. 3F; see Table S1E in the supplementary material). The reconstitution of the complete hypocotyl region apical to the GNOM expression domain is an impressive example of a GNOM-dependent effect that is locally separated from its zone of expression. Because GNOM expression in the hypophysis region is not required for restoring the root meristem (Fig. 2C,D), the restoration of the root meristem with the hypophyseal promoter is best explained by the non-cellautonomous effect of GNOM on the neighboring provascular tissue. To further analyze the non-cell-autonomous effects of GNOM expression in the N9312>>GNOM rescue embryos, we examined PIN1 localization at GNOM expression boundaries. Polar localization of PIN1 was detected directly above the GNOM expression domain in the provascular region (Fig. 3G,H). Concomitant with the polar localization of PIN1 in the reconstituted provasculature, auxin accumulation indicated by DR5rev::3xVenus-N7 reporter activity in the apical region of the embryo, which is never observed in wild type, was reduced (Fig. 3I). Because the N9312 promoter is exclusively expressed in the hypophysis and suspensor of globular embryos, reconstitution of PIN1 polarization is a non-cell-autonomous effect of GNOM expression in basal cells on the apically adjacent cells. Reconstitution of basal PIN4 expression in N9217>>GNOM and N9312>>GNOM rescue embryos Other PIN genes, PIN3, PIN4 and PIN7 are also expressed in the embryo (Friml et al., 2003). Besides PIN1, only PIN4 is expressed at globular stage in the basal region, whereas PIN7 expression is confined to the suspensor and PIN3 expression commences during the heart stage (Friml et al., 2002; Friml et al., 2003). Importantly, pin4 mutants show strong upregulation of DR5rev::PEH in inner pro-embryo cells, suggesting a major role for PIN4 in apical-basal auxin transport in the embryo (Friml et al., 2002). Therefore, we analyzed whether the localization pattern of PIN4 is also affected by restricted GNOM expression. As described by Friml et al. (Friml et al., 2002), PIN4 is expressed at the basal embryo pole in wild-type embryos and shows a rather unpolar distribution (Fig. 4A). Although PIN4 expression at the basal embryo pole was still observed in globular stage gnom embryos (Fig. 4B), PIN4 expression was strongly reduced by heart stage (Fig. 4C). However, basal PIN4 expression was maintained throughout heart stage in N9217>>GNOM rescue embryos (Fig. 4D) and at least partially in N9312>>GNOM rescue embryos (Fig. 4E). In conclusion, rescue of the primary root by GNOM expression in the provasculature or the hypophysis can be explained by the restoration of basal PIN1 localization and PIN4 expression in the provascular tissue and the root pole region, respectively, and a reconstitution of auxin transport towards the basal pole. Dose-dependent rescue activity of GNOM The ratio of the individual rescue phenotypes in the segregating populations differed, depending on the copy number of GNOM reporter transgenes: lines carrying only one copy of the driver construct (promoter::LhG4 or promoter::GAL4) and one copy of the reporter construct (pOp::GNOM or UAS::GNOM) showed a DEVELOPMENT Auxin transport and embryo patterning RESEARCH ARTICLE Fig. 4. Reconstitution of basal PIN4 expression in N9217>>GNOM and N9312>>GNOM gnom embryos. Immunofluorescence analysis of PIN4 (red) in wild-type heart stage embryo (A), and globular (B) and heart stage (C) gnom embryo. (D,E)Reconstituted PIN4 expression in N9217>>GNOM (D) and N9312>>GNOM (E) gnom embryo (torpedo stage) also carrying a GFP reporter. distinctive weaker rescue of gnom mutant seedlings than did homozygous lines carrying two copies of both constructs (compare Fig. 2C with 2D, see Table S1 in the supplementary material). Detailed analysis of the N9217>>GNOM and AtHB8>>GNOM rescue phenotypes in the F2 and F3 generations revealed a reduced number of fused cotyledons in favor of single monocot or dicot seedlings in homozygous lines (see Table S1 in the supplementary material): N9217>>GNOM homozygous lines displayed an increased number of monocot seedlings (Fig. 2D) and a simultaneous reduction in seedlings with fused cotyledons (Fig. 2C; see Table S1C in the supplementary material). Whereas hemizygous ATHB8>>GNOM lines only resulted in monocot rescue phenotypes (Fig. 2G), homozygous lines segregated a high percentage of rescued seedlings with two cotyledons that often appeared as nearly wild type (Fig. 2H, see Table S1D in the supplementary material). Because the radialized apex in gnom is associated with high apical auxin accumulation, indicated by DR5rev::GFP, the formation of one cotyledon could be the result of a restored apicalbasal auxin flux and reduced apical auxin accumulation. Indeed, hemizygous N9217 transactivation embryos that express the DR5rev::GFP marker apically develop fused cotyledons (Fig. 2C,S), whereas homozygous N9217 transactivation lines that show reduced DR5rev::GFP signals display an increase in monocot rescue phenotypes (Fig. 2D,T). Interestingly, although monocots were formed, even in homozygous N9217 rescue phenotypes the bilateral symmetry was not re-established. Auxin accumulation might therefore only partially explain the apical gnom phenotype. Development 138 (1) Radial-to-bilateral symmetry transition requires protodermal GNOM expression in the apical region To determine how GNOM might be involved in symmetry transition and organ formation, we expressed GNOM exclusively in the apical region of gnom mutant embryos using the AINTEGUMENTA (ANT) promoter (Fig. 5A). ANT>>GFP expression was initially observed in the protoderm of late globular/transition stage embryos. In contrast to wild-type embryos, where ANT>>GFP expression was subsequently restricted to the developing cotyledons (Fig. 1D), ANT>>GFP expression in gnom stayed confined to the protodermal layer (Fig. 5A). ANT>>GNOM rescue seedlings showed either fused cotyledons or a completely reconstituted apical pole with two cotyledons and a functional shoot meristem, which was GNOM dose dependent (Fig. 5C,D; see Table S1B in the supplementary material). In both cases, the cotyledons were expanded and showed a reconstituted vascular pattern (see Fig. S3B in the supplementary material), although the seedlings still lacked a primary root (Fig. 5D). In addition, ANT>>GNOM rescue seedlings that displayed bilateral symmetry also formed elongated primary leaves (Fig. 5D), which has never been observed for gnom, demonstrating the reconstituted activity of the shoot apical meristem. The fused apical phenotype was reminiscent of cuc1 cuc2 double mutants, which are impaired in organ boundary formation (Aida et al., 2002; Vroemen et al., 2003). Indeed, CUC2 localization was disrupted in gnom (not shown). Because the ANT promoter was predominantly expressed in the apical epidermis of gnom (Fig. 5A), epidermal expression of GNOM might be sufficient for the transition from radial to bilateral symmetry at the late-globular/transition stage. Although homozygous ANT>>GNOM rescue lines segregated an increased amount of offspring with a complete bilateral rescue, a significant proportion of rescued seedlings still displayed fused cotyledons, suggesting that the GNOM expression level or onset might be limiting (see Table S1B in the supplementary material). To further analyze the requirement for GNOM in the epidermis, we expressed GNOM from an epidermis-specific promoter in the gnom mutant background. The LIPID-TRANSFER-PROTEIN1 (LTP) promoter is exclusively expressed in the epidermis once the protoderm is formed (Thoma et al., 1994; Baroux et al., 2001). Driving GNOM from this promoter did not result in a discernible rescue phenotype, but introducing the LTP driver construct into the ANT>>GNOM background (LTP+ANT>>GNOM) led to a significant increase in the proportion of rescue phenotypes that showed a complete apical rescue with bilaterally symmetrical spacing of the cotyledons and a corresponding reduction of fusedcotyledon rescue phenotypes (see Table S1B in the supplementary Fig. 5. Rescue analysis of gnom mutants expressing GNOM from an apical promoter. Confocal and light microscopic analysis of ANT>>GNOM and ANT+LTP>>GNOM transactivation lines. (A)ANT>>GFP in gnom embryo (early heart stage). (B)ANT>>GFP in ANT>>GNOM gnom embryo (late heart stage). (C,D)ANT>>GNOM gnom segregating seedlings from lines expressing either one (C) or two (D) copies of ANT>>GNOM (representative phenotypes selected: see Table S1 in the supplementary material). (E,F)DR5rev::GFP in ANT>>GNOM gnom (E, globular stage; F, heart stage). (G)PIN1 localization (red) in ANT>>GNOM gnom early heart stage embryo also carrying a GFP reporter. (H)DR5rev::GFP in ANT+LTP>>GNOM gnom globular-stage embryo. Scale bar: 0.2 mm in C; 1 mm in D. DEVELOPMENT 122 material). This result demonstrates that epidermal GNOM expression can induce the radial-to-bilateral transition at the lateglobular/transition stage of embryogenesis. To determine whether the ANT>>GNOM rescue phenotype is linked to auxin, we analyzed the DR5rev::GFP expression pattern in the rescued mutant background. We still observed a strong apical DR5rev::GFP signal in globular-stage embryos (Fig. 5E), which became concentrated in the epidermal layer during later stages (Fig. 5F). In addition, ANT>>GNOM rescue embryos still showed a non-polar provascular PIN1 localization at heart stage (Fig. 5G), explaining the failure to rescue the primary root. By contrast, the LTP+ANT>>GNOM lines showed an increased proportion of embryos with a drastically reduced auxin level in the apical region (Fig. 5H). Altogether, these results strongly suggest that disrupted PIN1 localization in the provasculature inhibits auxin drainage from the apical pole of the globular gnom embryo, and the resulting apical auxin accumulation is likely to explain the observed organ fusion. Similar organ fusion events have been previously observed in shoot apical meristems treated with auxin (Reinhardt et al., 2000). GNOM expression in the provasculature or the hypophysis restores the basal ‘auxin sink’ (i.e. auxin drainage to the base), and single cotyledons (monocots) form in place of a radially fused apex. Finally, epidermal GNOM expression can transform otherwise fused cotyledon or monocot phenotypes to a wild-type appearance. Correct organ positioning therefore additionally requires epidermal GNOM. If epidermal GNOM levels are low (as in ANT>>GNOM), high apical auxin levels may still counteract correct organ positioning, explaining the observed fusion phenotypes. DISCUSSION Early embryo patterning requires inter-cellular communication events mediated by GNOM GNOM and GNL1 are redundantly required for ER-Golgi trafficking, precluding an embryo-lethal phenotype for each single mutant (Richter et al., 2007). Interestingly, gnom mutants still show a dramatic embryo patterning defect not seen for gnl1 or any other ARF GEF single knockout, owing to an additional role of GNOM in endosomal vesicle trafficking (Geldner at al., 2003). The latter function, however, is not required for cell survival, as gnom cell cultures are viable (Mayer et al., 1993). Our marker analysis revealed specific patterning defects in the apex and base of gnom mutant embryos. By contrast, radial tissue organization appeared unaffected (Fig. 1). This indicated a difference of GNOM requirements in individual cells of the embryo, despite the ubiquitous GNOM expression (see Fig. S1 in the supplementary material), and prompted us to do further research. Using tissuespecific GNOM expression, we show that apical and basal patterning of wild-type embryos depends on GNOM expression in the provasculature (Fig. 2). This non-cell-autonomous rescue of both the gnom embryo apex (partial rescue) and base was always accompanied by a non-cell-autonomous realignment of PIN1 in the non-GNOM expressing regions. Furthermore, central GNOM expression also induced non-cell-autonomous apical and basal auxin responses, as demonstrated by DR5rev::GFP and PIN4 localization studies. Wild-type DR5rev::GFP and PIN4 expression patterns were only observed after central or basal GNOM expression had reconstituted apical-basal PIN1 alignment. The results demonstrate that GNOM mediates intercellular signaling processes required for early embryo pattering and imply that the gnom phenotype is mainly caused by the unique function of GNOM in PIN1 recycling. RESEARCH ARTICLE 123 Fig. 6. Cell-autonomous and non-cell-autonomous GNOM function during embryogenesis. (A)Summary of GNOM requirement during embryogenesis. Vascular GNOM expression (green) is required to set up the root meristem at the early-globular stage (left). Epidermal GNOM expression (red) is required to set up and maintain epidermal auxin transport to cotyledon initiation sites at the lateglobular/transition stage (right). (B)Sink model for auxin transport in non-GNOM-expressing regions. GNOM expression in a basal domain (green) induces auxin drainage (blue arrows) from apical regions of gnom embryos [N9217>>GNOM (left) or N9312>>GNOM (right)]. GNOM-mediated auxin drainage induces polar PIN1 localization (red) also in neighboring tissues directly above the GNOM expression domain (non-cell-autonomous effect). GNOM mediates apical-basal auxin flux The initiation of the root meristem requires two or more apical MONOPTEROS (MP)-dependent signals that reach the uppermost suspensor cell during the early-globular stage to establish its hypophyseal cell fate and to trigger its asymmetric division (Weijers et al., 2006; Schlereth et al., 2010). One of these signals seems to be auxin, because disruption of the auxin response by inhibiting MP function in the embryo downregulates PIN1 expression and presumably limits auxin transport to the basal pole (Weijers et al., 2006; Ploense et al., 2009). Consistent with auxin being one of these signals, provascular GNOMdependent auxin transport is able to rescue primary root formation: provascular GNOM expression re-established provascular PIN1 polarity, reduced apical auxin accumulation and maintained PIN1 and PIN4 expression at the basal embryo pole (Fig. 2O; Fig. 4D,E; compare with Fig. 2J; Fig. 4C). As PIN1 and PIN4 are known to be induced by auxin (Friml et al., 2002; Vieten et al., 2005), the reconstitution of basal PIN4 expression is best explained by a reconstituted apical-basal auxin flow. Because radial patterning is not disrupted in gnom (compare Fig. 1I-K with 1M-O), the maintenance of embryo radial patterning seems to be independently controlled and not affected by disruption of apical-basal auxin flow. Given the implicated importance of PIN1 and PIN4 in embryonic patterning, one might expect severe apical and basal defects in pin1pin4 double-mutant embryos, considering their proposed role in apical-basal auxin flow in the embryo. However, this is not the case (Friml et al., 2003; Blilou et al., 2005), possibly because of compensatory expression patterns of other PIN family members (Vieten et al., 2005). By contrast, PIN recycling is DEVELOPMENT Auxin transport and embryo patterning RESEARCH ARTICLE blocked in gnom mutant embryos, regardless of compensatory PIN expression patterns, which accounts for the phenotypic similarity of gnom with pin1,3,4,7 quadruple knockouts. GNOM-dependent organ formation and symmetry transition in the embryo Provascular GNOM expression leads to a simultaneous reduction of apical organ fusion, linking gnom apical defects to the disruption of apical-basal auxin transport (Fig. 2C,D,S,T). Importantly, apical defects strongly depend on the dose of GNOM expression: increasing the expression level of GNOM in the central provascular tissue further reduces the apical DR5rev::GFP signal and induces a stronger non-cell-autonomous apical rescue. This correlates apical organ fusion directly with apical auxin content (Fig. 2C,D,S,T; see Table S1 in the supplementary material). Although GNOM expression in the provasculature reduced both apical auxin accumulation and apical fusion, only additional GNOM expression in the epidermis converted the radial symmetry of the globular embryo to the bilateral symmetry of the heart-stage embryo (Fig. 5D; see Table S1B in the supplementary material), arguing that epidermal auxin transport (towards the sites of cotyledon initiation) requires protodermal GNOM function (Fig. 6A). Similar to the GNOM function during lateral root initiation, where GNOM is required for PIN1 repolarization and the redirection of auxin flow (Geldner et al., 2004), epidermal GNOM expression in late-globular embryos may also reorient auxin flow for cotyledon initiation. The importance of the epidermis in cotyledon formation has already been implicated by several other studies (Abe et al., 2003; Benkova et al., 2003), but this study is the first direct link between epidermal auxin flow and cotyledon separation. Two promoters that are active in the provascular tissue, AtHB8 and N9217, differed in their ability to rescue the gnom mutant phenotype. The nearly complete rescue by AtHB8-driven GNOM expression might be related to the auxin-responsiveness of the AtHB8 promoter (Baima et al., 1995), which would confer additional GNOM expression in auxin-accumulating tissues. The AtHB8 promoter is indeed also expressed in protodermal tissue, unlike N9217 (compare Fig. 1L with 1K), which supports the notion that radial-to-bilateral symmetry transition requires protodermal GNOM expression. GNOM-induced auxin drainage GNOM vascular expression in cells above the hypophysis (N9217>>GNOM) rescued the root meristem region, hypocotyl elongation and cotyledon expansion (Fig. 2B,D). Whereas the effect on the root meristem and cell elongation along the axis can be explained by reconstituted auxin flow to the basal pole, the partial rescue of the apex seems to be caused by GNOM-mediated auxin drainage from the apical region (see Fig. 2L,S,T; Fig. 6B). In line with an apical auxin drainage induced by provascular GNOM expression, we observed a reconstitution of PIN1 polar localization in apical non-GNOM expressing regions (e.g. N9217>>GNOM, Fig. 2M-P). Similarly, hypophyseal GNOM expression resulted in re-establishment of the provascular PIN1 pattern and provascular auxin drainage (N9312>>GNOM, Fig. 3GI). In both cases, auxin drainage was driven by a basal ‘auxin sink’ formed by basally expressed GNOM. Consistent with basal ‘sink’ formation, PIN4 expression was upregulated upon provascular or hypophyseal GNOM expression. PIN4 has already been implicated in auxin sink and apical-basal auxin gradient formation, based on Development 138 (1) its localized expression pattern and its mutant phenotype (Friml et al., 2002). Similar sink-based auxin transport models have been suggested by the experimental work of Sachs and the computational modeling of auxin transport (Rolland-Lagan and Prusinkiewicz, 2005; Garnett et al., 2010). During leaf growth, for example, newly forming leaf veins originate from a distal auxin source near the leaf border, as well as a central auxin sink of already formed primary veins (Scarpella et al., 2006). Polar PIN1 targeting was previously thought to require GNOM in each individual cell but this had not been tested experimentally by generating genetic mosaics. By contrast, the non-cellautonomous GNOM function identified here suggests that cells that do not express GNOM still display PIN1 polarity possibly because of polar auxin flow towards the suggested GNOM-dependent basal auxin sink (Fig. 6B). In support of this idea, although individual cells of gnom mutant embryos can still retain PIN1 polarity, there is no coordinated polar localization of PIN1 between cells and relative to the apical-basal axis (Steinmann et al., 1999). How GNOM could establish an auxin sink is not at all clear but this appears to involve its role in polar recycling of PIN1 to the basal plasma membrane (Geldner et al., 2003). Further experiments will be required to address this issue. In principle, effects of a gene outside its expression domain can also be due to movement of the encoded protein or technical limitation of detection. Kim et al. (Kim et al., 2005), for example, described size restrictions for proteins to move within embryonic tissues. Being much larger than 3⫻GFP whose cell-to-cell movement was very limited, GNOM should be restricted to the cells of expression. This is also supported by the specificity of observed rescue phenotypes of different transactivation lines: although suspensor and globular embryo form a symplastic continuum for 1⫻GFP (27 kDa) (Stadler et al., 2005), GNOM transactivation from the suspensor and hypophyseal promoters showed distinct phenotypes. Detection of low-level protein expression is always problematic. However, we enhanced the sensitivity of signal detection by using GFP antibodies and signal amplification. Although some additional weak signals were detected at late stages of embryo development in some transactivation lines, no additional GFP expression was detectable in early stages when embryo patterning occurs. It is therefore unlikely that undetectable expression levels account for the noncell autonomous effects of GNOM. In summary, all the observed patterning defects in gnom could be linked to defects in embryonic auxin flow. GNOM requirements in the vasculature and the epidermis can be explained by GNOM requirements in vascular and epidermal polar auxin flow, respectively. Using tissue-specific GNOM expression, we were able to directly activate each one of these auxin transport routes individually, and analyze the resulting phenotype. We cannot exclude that GNOM-mediated vesicle traffic also affects other signaling pathways required for normal embryonic patterning, but it seems that all other signaling defects in gnom are masked by the auxin transport defect. In addition, our results suggest that the generation of auxin sinks may contribute to the regulation of plant patterning. Acknowledgements We thank M. Lenhard, A. Schlereth, D. Weijers, P. Brewer, Y. Eshed, S. Gordon, I. Moore, P. Gallois, R. Groß-Hardt, J. Friml, P. Benfey, K. Palme and the Nottingham Stock Centre for providing materials. We also thank C. Kägi, P. Sappl, M. Heisler and D. Weijers for critical reading and suggestions on the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (SFB 446, A9). DEVELOPMENT 124 Competing interests statement The authors declare no competing financial interests. Supplementary material Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.059147/-/DC1 References Abe, M., Katsumata, H., Komeda, Y. and Takahashi, T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635-643. Aida, M., Vernoux, T., Furutani, M., Traas, J. and Tasaka, M. (2002). Roles of PINFORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965-3974. Alvarez, J. P., Pekker, I., Goldshmidt, A., Blum, E., Amsellema, Z. and Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18, 1134-1151. Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I. and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171-4182. Baroux, C., Blanvillain, R., Moore, I. R. and Gallois, P. (2001). Transactivation of BARNASE under the AtLTP1 promoter affects the basal pole of the embryo and shoot development of the adult plant in Arabidopsis. Plant J. 28, 503-515. Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jürgens, G. and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591-602. Berleth, T. and Juergens, G. (1993). The role of the monopteros gene in organising basal development in the Arabidopsis embryo. Development 118, 575-587. Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K. and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 3944. Breuninger, H., Rikirsch, E., Hermann, M., Ueda, M. and Laux, T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867-876. Busch, M., Mayer, U. and Jürgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 250, 681-691. Casanova, J. E. (2007). Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8, 1476-1485. Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M. G., Feldmann, K. A. and Benfey, P. N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423-433. Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G. et al. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661-673. Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R. and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 413, 147-153. Garnett, P., Steinacher, A., Stepney, S., Clayton, R. and Leyser, O. (2010). Computer simulation: the imaginary friend of auxin transport biology. BioEssays 32, 828-835. Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A. and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediatesendosomal recycling, auxin transport, and auxindependent plant growth. Cell 112, 219-230. Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R. A., Mayer, U. and Jürgens, G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131, 389-400. Hadfi, K., Speth, V. and Neuhaus, G. (1998). Auxin induced developmental patterns in Brassica juncea embryos. Development 125, 879-887. Hamann, T., Mayer, U. and Jürgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387-1395. Hamann, T., Benkova, E., Bäurle, I., Kientz, M. and Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610-1615. Hardtke, C. S., Ckurshumova, W., Vidaurre, D. P., Singh, S. A., Stamatiou, G., Tiwari, S. B., Hagen, G., Guilfoyle, T. J. and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors RESEARCH ARTICLE 125 MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089-1100. Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58, 139-151. Heisler, M. G., Ohno, C., Das, P., Sieber, P., Reddy, G. V., Long, J. A. and Meyerowitz, E. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899-1911. Jürgens, G. and Mayer, U. (1994). Arabidopsis. In EMBRYOS. Colour Atlas of Development (ed. J. B. L. Bard), pp. 7-21. London: Wolfe Publishing. Jürgens, G., Mayer, U., Torres Ruiz, R. A., Berleth, T. and Misera, S. (1991). Genetic analysis of pattern formation in the Arabidopsis embryo. Development 1, 27-38. Kim, I., Kobayashi, K., Cho, E. and Zambryski, P. C. (2005). Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102, 11945-11950. Laplaze, L., Parizot, B., Baker, A., Ricaud, L., Martinière, A., Auguy, F., Franche, C., Nussaume, L., Bogusz, D. and Haseloff, J. (2005). GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56, 2433-2442. Lauber, M. H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W. and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485-1493. Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., Alvarez, J. P. and Eshed, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103, 6398-6403. Long, J. A. and Barton, M. K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027-3035. Mayer, U., Büttner, G. and Jürgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development 117, 149-162. Moore, I., Gälweiler, L., Grosskopf, D., Schell, J. and Palme, K. (1998). A transcription activation system for regulated gene expresssion in transgenic plants. Proc. Natl. Acad. Sci. USA 95, 376-381. Müller, B. and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094-1097. Nakajima, K., Sena, G., Nawy, T. and Benfey, P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307-311. Ploense, S. E., Wu, M. F., Nagpal, P. and Reed, J. W. (2009). A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136, 1509-1517. Reinhardt, D., Mandel, T. and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507-518. Richter, S., Geldner, N., Schrader, J., Wolters, H., Stierhof, Y.-D., Rios, G., Koncz, C., Robinson, D. G. and Jürgens, G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448, 488492. Rolland-Lagan, A. G. and Prusinkiewicz, P. (2005). Reviewing models of auxin canalization in the context of leaf vein pattern formation in Arabidopsis. Plant J. 44, 854-865. Sabatini, S., Heidstra, R., Wildwater, M. and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354-358. Scarpella, E., Marcos, D., Friml, J. and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015-1027. Scheres, B., Di Laurenzio, L., Willemsen, V., Hauser, M. T., Janmaat, K., Weisbeek, P. and Benfey, P. N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121, 53-62. Schlereth, A., Möller, B., Liu, W., Kientz, M., Flipse, J., Rademacher, E. H., Schmid, M., Jürgens, G. and Weijers, D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913-916. Scholl, R. L., May, S. T. and Ware, D. H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124, 1477-1480. Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G. and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635644. Shevell, D. E., Leu, W. M., Gillmor, C. S., Xia, G., Feldmann, K. A. and Chua, N. H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051-1062. Stadler, R., Lauterbach, C. and Sauer, N. (2005). Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 139, 701-712. DEVELOPMENT Auxin transport and embryo patterning RESEARCH ARTICLE Steinmann, T., Gelder, N., Grebe, M., Mangold, S., Jackson, C. L., Paris, S., Gälweiler, L., Palme, K. and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316318. Takada, S. and Jürgens, G. (2007). Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141-1150. Thoma, S., Hecht, U., Kippers, A., Botella, J., De Vries, S. and Somerville, C. (1994). Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 105, 35-45. Vieten, A., Vanneste, S., Wisniewska, J., Benkova, E., Benjamins, R., Beeckman, T., Luschnig, C. and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521-4531. Development 138 (1) Vroemen, C. W., Mordhorst, A. P., Albrecht, C., Kwaaitaal, M. A. and De Vries, S. C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563-1577. Weijers, D., Franke-van Dijk, M., Vencken, R. J., Quint, A., Hooykaas, P. and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semidominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289-4299. Weijers, D., Schlereth, A., Ehrismann, J. S., Schwank, G., Kientz, M. and Jürgens, G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265-270. Wysocka-Diller, J. W., Helariutta, Y., Fukaki, H., Malamy, J. E. and Benfey, P. N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595-603. DEVELOPMENT 126