* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download HLA-A, -B
Zinc finger nuclease wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA paternity testing wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Designer baby wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
SNP genotyping wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Microevolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Helitron (biology) wikipedia , lookup
HLA A1-B8-DR3-DQ2 wikipedia , lookup
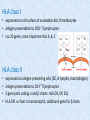
Donor Selection for HSCT Prof. Ilona Hromadníková, Ph.D. Department of Molecular Biology and Cell Pathology Third Medical Faculty, Charles University in Prague Selection criteria • HLA compatibility If selection is possible among more donors consideration of: • age younger is more suitable, usually not donors > 60 years Czech Donor Register: Donor age has to be between 18 – 35 years when firstly included into the registr. • sex female to male BMT – risk of GvHD especially when female is alloimmunized against H-Y and other antigens of minor histocomp. system, male to female with SAA BMT – higher rejection risk Selection criteria If selection is possible among more donors consideration of : • AB0- and Rh- blood groups not significant risk factor, however, necessary to know if incompatible – remove erythrocytes and plasma • immunity against CMV risk of infection transfer, when donor pos. and recipient neg. CMV positivity → unfavourable influence on development and course of GvHD even if no clinical symptoms of infection Contra-indication for donation • infectious diseases transmitted by blood antibody screening: syphilis, hepatitis B, C, HIV-1/HIV-2, CMV (IgM, IgG) • because of general anaesthesia mainly: cerebrovascular diseases fresh heart attack respiratory insufficiency malignancies pregnancy HLA system of histocompatibility • huge gene polymorphism • Tx - important antigens HLA class I A, B, C HLA class II DR, DP, DQ HLA class I • expressed on cell surface of nucleated cells, thrombocytes • antigen presentation to CD8+ T lymphocytes • cca 20 genes, most important HLA A, B, C HLA class II • • • • expressed on antigen presenting cells (DC, B lympho, macrophages) antigen presentation to CD4+ T lymphocytes 3 gene pairs coding a and b chains: HLA DR, DP, DQ HLA DR: a chain is monomorphic, additional gene for b chain Polymorphism HLA locus HLA-A HLA-B HLA-C HLA-DR (only b chain) HLA-DQ (a and b chains) HLA-DP (a and b chains) Antigenic variants 25 53 11 20 9 6 DNA variants 83 186 42 221 49 88 Clinical genetics, 2001 antigenic variants (specificities) – due to differences in amino acid composition in a or b chain, determined by serology DNA variants – in HLA alleles defined by serology were found further variations in DNA sequence Example: HLA B27 allele is serologically unique, however 12 different variations in nucleotide sequence were found Nomenclature of HLA system HLA A*0101 locus serologic specificity specific allele defined by nucleotide sequencing Example: HLA A*0101, HLA A*0102 – same serologic specificity A1 differing in nucleotide sequence HLA DRB1*0401 class subtype serologic specificity chain a/b number marking particular gene specific allele labelling w = working labelling Polymorphism and inheritance of HLA haplotypes • high polymorphism number (not all!) causes variability in structure of surface HLA proteins • HLA alleles transmitted together like haplotypes • each parent has 2 expressed haplotypes child will have one or another → 25% chance exists, that two siblings will have identical HLA haplotype as one of the parents AB + CD → AC / AD / BC / BD • high variability in profile and frequence of HLA variants example: HLA A2 most frequent in all populations HLA A24 in Caucasians, not in afro-Americans and Asians • ethnically different distribution of particular HLA alleles and haplotypes Incompatibility in Tx can lead to: • graft failure role of HLA-A, -B, -C, -DR mismatches, total number of disparities influences the risk of graft failure • GvHD development mismatches in HLA class I and/or II increase risk of aGvHD • decreased survival mismatches in HLA-A, -B, -C, -DR but not -DQ, -DP decrease survival according to U.S. study outcome differs according to individual studies HLA typing Serotyping • identification of HLA class I and II • discrimination of protein molecules on the basis of diversity in antigenic characteristics • using typing antisera panel (commercial trays), blood of multiparous women (1 father) – serum contains sufficient amount of antibodies against HLA molecules of foetus inherited from the father • very laborious procedure • less time-consuming than genotyping • certain degree of inaccuracy HLA typing Serotyping • Routine typing of HLA class I: 120 typing sera in microtitration plate tested T lymphocytes are added, after incubation complement is added positivity = serum antibodies bind to T lymphocytes, lysed by complement evaluation using fluorescent microscope (dye binding to DNA in lysed cell) • Routine typing of HLA class II: 60 typing sera in panel tested B lymphocytes are added, microcytotoxic test HLA DR, DQ testing HLA genotyping DNA (genotyping) • using PCR and specific primers for individual allele, DNA isolation from blood • 2 approaches: low resolution – identification of broad families of alleles that cluster into serotypes („2-digit typing“, e.g. A*02 = A2 in serology) high resolution – identification of the individual alleles within each serotype („4digit typing“, e.g. A*0201) • more time-consuming • simple method, easy automatization • more sensitive method Example: serotyping defines A*02, another 64 known alleles (A*0201-0264) genotyping defines A*0201 allele Clinical scheduling usually: HLA class I serotyping, HLA class II genotyping HLA typing - example Serotyping defines individual serotype: A1,A3,B7,B8,DR3,DR15(2),DQ2,DQ6(1) Genotyping specifies HLA phenotype in individual: A*0101, *0301, Cw*0701,*0702, B*0702,*0801, DRB1*0301,*1501, DQA1*0501,*0102, DQB1*0201,*0602 composed from 2 haplotypes from the parents: A*0101 : Cw*0701 : B*0801 : DRB1*0301 : DQA1*0501 : DQB1*0201 (by serotyping A1-Cw7-B8-DR3-DQ2) A*0301 : Cw*0702 : B*0702 : DRB1*1501 : DQA1*0102 : DQB1*0602 (by serotyping A3-Cw7-B7-DR15-DQ6) HLA typing of blood relatives Typing of patient and related donor • HLA-A, -B, -DR typing of two digits behind * mostly sufficient for identification of maternal and paternal haplotypes • confirmation of genotypic identity for the whole set of HLA genes (A,B,C,DR,DP,DQ) on both chromosomes = match 12/12 • HLA-DP usually not tested – match 10/10 HLA typing of blood relatives IHBT, National reference laboratory for DNA diagnostics Department of HLA analysis • genotyping always with indication for HSCT, concurrently blood taking from primary blood relatives (siblings, parents, eventually children of the patient) • standard typing of HLA-A, -B, -DRB1 on the level of allele groups (low resolution) and identification of both haplotypes = serotyping of HLA class I and genotyping of HLA class II - „2digits“ typing • unclarity in haplotype identification: genotyping of individual alleles (high resolution, „4-digits“ typing) Primary sample submission form for HLA genotyping genotypizaci After determination of match/mismatch with relatives: a) HLA-identical related donor in close family found (i.e. sibling, parent) blood taking for confirmatory examination, low resolution genotyping in loci: HLA-A,-B,-C,-DRB1,-DQB1 (prior to starting conditioning regimen before Tx) a) HLA-identical related donor in close family not found searching in extended family - uncle, cousin... (HLA-A,-B,-DRB1 low resolution) or indication for unrelated HSCT (registry searching) after finding the donor: confirmatory examination by high resolution genotyping of loci: HLA-A,-B,-Cw,-DRB1,-DQB1 HLA typing of unrelated donors • has to be typed: HLA- A*,B,Cw* , DRB1* a DQB1* • matching degree expressed as a 10/10, 9/10, 8/10 match • optional examination of genes DPB1*, DRB3*-5* or DQA1* when several matched donors identified (also match in: AB0, age, sex, CMV status) • match evaluated according to results of „4-digits" typing pair - DRB1*1101 vs. DRB1*1103 mismatched • HLA mismatch preferences for possible donor choice: Cw*>A*B*,DQB1*>DRB1 Cw* the most accepted mismatch, DRB1 least accepted mismatch • non-HLA mismatches (blood group, CMV) can influence the choice of the donor more than HLA mismatch hierarchy, see the example: HLA typing of unrelated donors Example patient: A*0201, B*3501,1501, Cw* 0401,0303… CMVneg, BG Aneg donor #1: A*0201, B*3503, 1501, Cw*0401, 0303, CMVneg , BG Aneg donor #2: A*0201, B*3501, 1501, Cw*0401, 0304, CMVpos, BG Bpos -> donor #1 is preferred despite of HLA-B gene mismatch (CMV +ABO compatibility) Tx from HLA-identical sibling Patient: M/4, MDS graft: BM donor: sister Blood group HLA Match 10/10 weight: 36 kg age: 9 Recipient 0 Rh + Donor 0 Rh + A *0201 *6801 A dtto B *3501 *3906 B dtto Cw *0401/04 * 1203 Cw dtto DRB1 *0101 *0801 DRB1 dtto DQB1 *0501 *0402 DQB1 dtto DPB1 DPB1 dtto Tx from unrelated donor Patient: M/16, pre-B ALL CR2 graft: PBSC Donor code: DERKS 900111472 Sex: F weight: 87 Blood group HLA Match 7/10 A Recipient Rh age: 40 - A Donor Rh + A B Cw DRB1 *0201 *2705 *0202 *0101 *2402 *4101 *1602 *0404 A B Cw DRB1 *0201 *2705 *02 *0101/17-19 *0301 *4101 *17 *0404 DQB1 *0501 *0302 DQB1 *0501 *04 DPB1 * * DPB1 * * Searching in donor registries Czech republic has 3 registries • Czech Stem Cell Registry (IKEM, Prague) • Cord Blood Bank (IHBT, Prague) • Czech National Marrow Donor Registry (CNMDR, Pilsen) - established in 1992 in Pilsen by Bone Marrow Transplant Foundation, 7 donor centers created in CR 1993 cooperation via Bone Marrow Donor Worldwide located in Leiden, Netherlands 1997 cooperation contract with the American National Marrow Donor Program until 1998 funded entirely by Bone Marrow Transplant Foundation 2000 concluded mutual contracts with health insurance companies – cover some of the expenses in conjunction with active search for the most suitable donors from the Registry as well as the expenses connected with their more detailed HLA examination Donor centres in CR http://www.kostnidren.cz/registr/ http://www.czechbmd.cz/ • Brno • České Budějovice • Hradec Králové • Most • Olomouc • Ostrava • Plzeň • Praha • Ústí nad Labem cooperation with other donor centres in regions Global Donor Registry • Bone Marrow Donor Worldwide 14,093,962 (13,684,277 donors and 409,685 CBU's) (last updated: 22-Feb-2010) 60 bone marrow registries from 44 states + 42 cord blood registries from 26 states Germany cca 3,7 million donors, CR cca 55 000 USA – 14 registries, cca 5,5 million donors Anthony Nolan Trust • first bone marrow donors registry • established in 1974 in Great Britain (Westminster Children‘s Hospital) • by Anthony Nolan‘s mother (WAS, 1971 – 1979) • presently one of the biggest registries in the world • http://www.anthonynolan.org.uk/