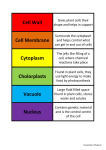
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Direct Drug Metabolism Monitoring in a Live Single Hepatic Cell by
Survey
Document related concepts
Cytoplasmic streaming wikipedia , lookup
Cell growth wikipedia , lookup
Cell nucleus wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Endomembrane system wikipedia , lookup
Cell culture wikipedia , lookup
Cytokinesis wikipedia , lookup
List of types of proteins wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Transcript
ANALYTICAL SCIENCES MARCH 2012, VOL. 28 201 2012 © The Japan Society for Analytical Chemistry Rapid Communications Direct Drug Metabolism Monitoring in a Live Single Hepatic Cell by Video Mass Spectrometry Sachiko DATE,* Hajime MIZUNO,** Naohiro TSUYAMA,** Takanori HARADA,** and Tsutomu MASUJIMA*,**† *Quantitative Biology Center (QBiC), RIKEN, 6-2-3 Furuedai, Suita, Osaka 565–0874, Japan **Graduate School of Biomedical Sciences, Hiroshima University, 1-2-3 Kasumi, Minami, Hiroshima 734–8551, Japan The metabolism of anti-breast cancer drug, tamoxifen, in a single human hepatocellular carcinoma cell, HepG2, was directly monitored by a video-mass spectroscope. The cytoplasm, a vacuole or nucleus of the cell was directly sucked by a nano-spray tip under a video-microscope, and then was introduced into a mass spectrometer. Unchanged drug molecules were found in cytoplasm and a vacuole, but the metabolites were only found in the cytoplasm. This direct detection of drug metabolites in a live single cell is useful for speedy drug metabolism monitoring. (Received February 4, 2012; Accepted February 13, 2012; Published March 10, 2012) The monitoring of drug metabolism in a hepatic cell, or in a target cell is a very important trace analysis in drug discovery and development. However, these analyses have been mostly performed by liquid chromatography–mass spectrometry (LC-MS), which needs more than thousand cells. Cells were pretreated by sonication or homogeneization by which any information of drug localization within a cell was lost, in most cases. If the drug metabolism and the localization of drug metabolites within a live single cell can be directly observed under a video microscope, the drug discovery and development become speedy and precise. Furthermore, in personalized medicine,1 a drug can be properly selected by single-cell drug metabolism analysis using only a few cells in biopsied tissues. We developed the live single-cell video mass spectrometry,2–4 and applied to the analysis of live single-cell drug metabolism using a hepatic model cell, HepG2. HepG2 cells (human hepatocellular carcinoma cells) were cultured in 35 mm dishes at 37° C in 95% air/5% CO2 in Dulbecco’s Modified Eagle’s Medium with 4500 mg/L glucose, supplemented with 10% fetal bovine serum (FBS). FBS was inactivated by a heating treatment at 56° C for 30 min. Tamoxifen (TAM) was added to cultured HepG2 cells to be 5 mM final concentration, and incubated for 12 h. An organelle (cytoplasm, a vacuole or a nucleus) was sucked into a nanospray tip (HUMANIX) independently, as shown in Fig. 1. Then, 3 μL of a solvent (90% MeOH/8.0% H2O/2.0% formic acid) was added from the back-end of the tip. These samples were directly introduced by nano-electrospray ionization into a LTQ Orbitrap XL (Thermo Fisher Scientific) at a positive ion mode. Each metabolite was identified according to its exact mass m/z value of Δppm <±3.0. Tamoxifen (m/z 372.232) was detected in cytoplasm (Fig. 2). Its five metabolites,1,5–10 (N-desmethyl tamoxifen (N-DM-T, m/z 358,216), 4-hydroxy To whom correspondence should be addressed. E-mail: [email protected] † tamoxifen (4-OHT), tamoxifen-N-oxide (T-NO) (m/z 388.227), 3,4-dihydroxy tamoxifen (3,4-diOHT), 4-hydroxy tamoxifen N-oxide (4-OHT-NO) (m/z 404.222)) were also detected (Fig. 2). N-DM-T, which is mainly metabolized by CYP3A4, was preferably detected. This result is in accord with the finding that N-DM-T is a major metabolite of TAM,5 and that CYP3A4 is the most expressed isozyme in P450 subfamilies of the HepG2 cell.11 Weak molecular peaks were also identified by LC-MS/MS with many cells. Besides the peaks of tamoxifen and its metabolites, many peaks were detected at the same time, most of which were found in the control (without tamoxifen). Most of these peaks were hard to identify, however, peak origin was told apart. The peak of m/z 372.186, for example, is solvent peak, and the peak of m/z 372.267 is from cytoplasm. Two metabolites, (4-OHT and T-NO, which have the same mass (m/z 388.227), and two metabolites, 3,4-diOHT and 4-OHT-NO, that have the same mass (m/z 404.222)) were identified using LC-MS/MS methods, however, endoxifen which is known as the most active metabolites, was not detected. Since HepG2 is reported to have less metabolizing activity,11 these secondary metabolites can be found in greater numbers in primary cultured cells. In vacuoles, tamoxifen was detected (Fig. 3), but no metabolites were detected. On the other hand, neither tamoxifen nor metabolites were detected in the nucleus. These data indicate that trapped cytoplasm includes a metabolizing organelle, such as endoplasmic reticulum. Since tamoxifen metabolites were only found in cytoplasm, transport of metabolites into a vacuole or a nucleus seems to be minimal. The role of vacuole in the HepG2 cell is still unknown, however, the vacuole appears to be vacant and having no granules in it. Vacuole seems to be a reservoir or wasting envelope, and to have a less viscous aqueous solution, because it can be sucked easily and absorbed into the glass capillary inner wall when it is trapped. This study of live single-cell level drug metabolism analysis will open the possibility of speedy, low-cost and site-specific 202 ANALYTICAL SCIENCES MARCH 2012, VOL. 28 Fig. 1 Trapping of the cell contents of cytoplasm, a vacuole and a nucleus by a nanospray tip from a single HepG2 cell under a video microscope. monitoring of drug metabolism and a useful tool for single-cell tailored medicine using biopsied cells. Acknowledgements This work was supported by the Grant-in-Aid (No. 20229003) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. References 1. J. M. Hoskins, L. A. Carey, and H. L. McLeod, Nat. Rev. Cancer, 2009, 9, 576. 2. T. Masujima, Anal. Chim. Acta, 1999, 400, 33. 3. H. Mizuno, N. Tsuyama, T. Harada, and T. Masujima, J. Mass Spectrom., 2008, 43 ,1692. 4. T. Masujima, Anal. Sci., 2009, 25, 953. 5. G. K. Poon, B. Walter, P. E. Lønning, M. N. Horton, and ANALYTICAL SCIENCES MARCH 2012, VOL. 28 203 Fig. 3 Detected mass spectra for a vacucole from a single HepG2 cell that shows only TAM. R. McCague, Drug Metab. Dispos., 1995, 23, 377. 6. P. W. Fan, F. Zhang, and J. L. Bolton, Chem. Res. Toxicol., 2000, 13, 45. 7. F. Zhang, P. W. Fan, X. Liu, L. Shen, B. B. van Breeman, and J. L. Bolton, Chem. Res. Toxicol., 2000, 13, 53. 8. T. Kaku, K. Ogura, T. Nishiyama, T. Ohnuma, K. Muro, and A. Hiratsuka, Biochem. Pharmacol., 2004, 67, 2093. 9. Y. Zheng, D. Sun, A. K. Sharma, G. Chen, S. Amin, and P. Lazarus, Drug Metab. Dispos., 2007, 35, 1942. 10. R. S. Sane, D. J. Buckley, A. R. Buckley, S. C. Nallani, and P. B. Desai, Drug Metab. Dispos., 2008, 36, 946. 11. S. Wilkening, F. Stahl, and A. Bader, Drug Metab. Dispos., 2003, 31, 1035. Fig. 2 Detected mass spectra for cytoplasm from a single HepG2 cell that shows TAM and its metabolites.