* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Radioisotopes in biology
Survey
Document related concepts
Molecular evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Non-coding DNA wikipedia , lookup
List of types of proteins wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Molecular cloning wikipedia , lookup
Protein adsorption wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Proteolysis wikipedia , lookup
Point mutation wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
DNA vaccination wikipedia , lookup
Transcript
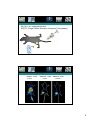
Radioisotopes in biology Particles produced by radioactive decay 1 Types of radioactive decay -α-particle emission (usually elements with high atomic number): He2+ emitted -positron emission (conversion of a proton to a neutron): proton Æ neutron + β+ -neutron emission (conversion of a neutron to a proton): neutron Æ proton + β-X-ray emission (fusion of proton and electron): proton + electron Æ neutron + X-ray -γ-ray emission does not change the atomic number or mass. It usually accompanies the release of an α- or β- particle that leaves the atom in an excited state. The atom can then reach a ground state by releasing a γ-particle. Radioactive decay energy -Radioactive decay energy is measured in eV -Definition: 1 eV is the energy absorbed by an electron in accelerating through a potential difference of 1V. α-particle (4-8 meV) γ-rays and β-particle (usually less than 3 MeV) 2 Half-lives of some isotopes used in biological studies Detection and quantification of radioactivity -ionization of gases -excitation of solids or solutions -exposure of photographic emulsions 3 Detection of radioactivity (by gas ionization) 1. A voltage is applied between the anode and cathode. 2. When a charged particle passing through the gas in the chamber it ionizes particles. 3. A current is formed which is measured. 4. Can be used to measure α- and β- particles but not γ-rays. Geiger-Müller tube Townsend avalanche effect Detection of radioactivity by excitation -Liquid scintillation counting -PM photomultiplier tube Solvent e.g. toluene Primary flour e.g. PPO= 2,5-diphenyl-oxazole Secondary flour e.g. POPOP=1,4-bis(5-phenyloxazol-2-yl)benzene 4 Working with radioactivity Sheilding suitable for work with β-emitters Geiger-müller counter Scintillation proximity assay (SPA) + -High throughput screening -Easy to automate -Versatile method 5 Exposure of photographic emulsions -Autoradiography: To locate the position of a radiation source within a sample. The sample is placed on a photographic emulsion and an image is produced much as a normal photo. Weak β-emittors are sutiable (e.g. 3H, 35S) -Very sentitive method, exposure for days or longer Tracing proteins in cells EXAMPLE: a pulse chase experiment 1. PULSE Grow cells in medium containing radioisotope for a period of time (in this example we use 35S). 2. CHASE: Replace hot (radio-isotope containing) medium with cold medium. 6 Pulse chase Example: 1. Take samples after certain times and isolate a desired biomolecule. 2. Quantify its amount of radioactivity One type of antibody fragment (diabody) Another type of antibody fragment (single chain) Tracing proteins in multicellular organisms Example: Tracing tumor cells by SPECT imaging 7 QuickTime™ and a GIF decompressor are needed to see this picture. In Vivo Targeting–Experimental Set-up–Professor Stahl. SK-OV-3 s.c. xenograft tumors SPECT (Single Photon Emission Computed Tomography) 125I QuickTime™ and a GIF decompressor are needed to see this picture. HER2-targeting - effect of affinity ZHER2:4 - 50 nM 6 hours a ZHER2:342 - 22 pM 6 hours b ZHER2:342 - 22 pM 24 hours c 8 QuickTime™ and a GIF decompressor are needed to see this picture. Biodistribution in mice Biodistribution (125I, n=4, 12h p.i.) brain bone muscle skin tumor thyroid/organ saliv gland larg int sm int stomach pancreas spleen liver lung heart 0 kidney 5 blood %ID/g 10 Protein-ligand interactions -interactions are influenced by physical parameters such as pH, temperature and ionic concentration. -It is important to allow the system to reach equilibrium The dissociation constant Kd for a particular interaction can be determined experimentally, through e.g. a scatchard plot. 9 Equlibrium dialysis Surface plasmon resonance (SPR) spectroscopy 10 Coupling to of ligands to the sensorchip Association and dissociation 11 Quartz crystal microbalance (QCM) biosensor Resonance frequency is changing as a result of mass deposition on the surface Electrophoretic mobility shift assay (EMSA) To analyze the interaction between protein and DNA/RNA 1. Radio-label the DNA (typically using 32P), but e.g. fluorescent probe is also ok. 2. Mix a fixed amount of DNA with a varying amount of protein 3. Separate the Protein DNA mixtures by SDS-PAGE. 12 EMSA DNA/Protein complex DNA Kd can be directly determined when half the DNA molecules are bound to Protein 13