* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Questions chapter 15
Cell-penetrating peptide wikipedia , lookup
Polyadenylation wikipedia , lookup
Molecular evolution wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein adsorption wikipedia , lookup
Transcriptional regulation wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Bottromycin wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
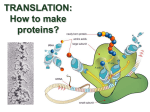
Chapter 15 Translation a. What is the Shine-Dalgarno sequence? What binds to the sequence during translation? Describe some of the ways in which the sequence can vary, addressing the consequences that any such variation has on translation efficiency. b. What is the role of the 5′ cap during eukaryotic translation? What happens to mRNA molecules that are missing a cap? c. Describe the structural and sequence elements that are common to all tRNA molecules, addressing the function of each of the elements. What forces stabilize the tRNAs' structural features? d. Outline the steps by which aminoacyl tRNA synthetases charge tRNAs. How can some organisms get away with having fewer than 20 synthetases, yet still charge tRNAs with all 20 amino acids? e. Outline the steps of the ribosome cycle. At what stage do the ribosomal subunits bind to each other? To mRNA? What causes them to dissociate when protein synthesis is complete? f. Describe the A, P, and E sites of the ribosome. Where are they located? What occupies each of the sites during the following stages of translation: • upon creation of the 70s initiation complex • after an initial round of peptidyl transfer, prior to aminoacyl tRNA binding • following aminoacyl tRNA binding, but before the peptidyl transfer reaction • after peptidyl transfer, but before translocation is completed by EF-G • in the presence of puromycin, prior to peptidyl transfer • upon reaching a stop codon • following release of the polypeptide chain, but before the appearance of RRF g. Describe the different mechanisms used to ensure translation accuracy, beginning with the charging of the tRNA and continuing up to the transfer of the amino acid to the growing polypeptide chain. How does the degree of translation accuracy compare to that of DNA replication or of transcription? Does it make sense that the rate should be lower for translation than it is for these other processes? h. How do prokaryotic cells recognize the stop codon during translation? How does recognition of the codon lead to the termination of protein synthesis and dissociation of the ribosomal subunits? What happens if the stop codon is missing? 1