* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene Regulation
Protein moonlighting wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Genome (book) wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic code wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Transcription factor wikipedia , lookup
Gene expression profiling wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Oncogenomics wikipedia , lookup
Microevolution wikipedia , lookup
Frameshift mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
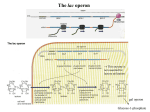
Gene Regulation I. Transcriptional Regulation Operons (clustering by function) regulatory region of an operon promoter operator leader region Negative regulation repressor proteins (may bind corepressor molecules) Positive regulation activator protein (may bind coactivator molecules) II. The lac Operon (Negative Regulation) Jacob and Monod proposed the operon model lacI, operator, promoter, lacZYA LacI, when bound to allolactose, binds operator to inhibit transcription initiation Isolation of Common Mutants lac- mutants could not grow on lactose lacc mutants lac genes were always expressed, whether lactose was present or not Defining the Operon (complementation tests) common recessive lac- mutations that complements other lac- mutations... ...gene products act in trans defines structural genes (such as lacZ-, lacY-) rare recessive lac- mutations that don't complement any complementation group... ...probably act in cis and could be a... -mutation in the promoter/operator region -polar mutation in first gene of the operon rare dominant lac- mutations... ...all map to lacI called lacIs (superrepressor) common recessive lacc mutations... ...all mapped to lacI inactivated lacIrare dominant lacc mutations... ...all mapped to lacI inactived LacI protein but it could still form tetramers As a Tool in Molecular Biology lac promoter is inducible. Allowing production of toxic genes IPTG, nonclevable derivative of allolactose Several colorimetric substrates exist for the lacZ gene product, beta-galactosidase ONPG turns yellow in the presence of beta-galactosidase X-gal turns blue in the presence of beta-galactosidase III. The L-ara Operon (Positive Regulation) Operon Structure araC, operators, inducers, araBAD Isolation of Mutants ara- mutants are still common but.. arac muants are rare because the mutation must make AraC active without binding arabinose Inactivation of araC (unlike lacI) produces an ara- phenotype AraC must also be an antiactivator since... araCc mutations should be dominant (but they are not). IV. The trp operon (Negative regulation and transcriptional attenuation) Negative regulation by trpR gene product TrpR only binds/represses operon when binding tryptophan... ...so TrpR is an aporepressor Transcriptional attenuation Leader sequence encodes a 14 amino acid leader polypeptide not a 'functional' protein, but acts to regulate transcription 2 trp codons at position 10 11 are key Four segments in the leader RNA that can form secondary structures Bottom Line of regulation if segments 3 and 4 form a hairpin. a transcriptional terminator is formed... ...and transcription terminates if segments 2 and 3 form a hairpin, the transcriptional terminator cannot ...form and transcription will continue The result is that tryptophan levels are regulated as a gradient rather than just on or off





















![[001-072] pierce student man](http://s1.studyres.com/store/data/017104672_1-497120343b128453965f3c5043e36ba3-150x150.png)
