* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download A Truncated Laminin Chain Homologous to the B2 Chain: Structure
Survey
Document related concepts
Proteolysis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gene regulatory network wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Expression vector wikipedia , lookup
Gene expression wikipedia , lookup
Point mutation wikipedia , lookup
Biosynthesis wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemistry wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcript
Published November 1, 1992
A Truncated Laminin Chain Homologous to the B2 Chain:
Structure, Spatial Expression, and Chromosomal Assignment
Pekka Kallunki,* Kirsi Sainio,r Roger Eddy,w M a r y Byers,w T u u l a Kallunki,* H a n n u Sariola,+
K o n r a d Beck,ll H a r r i Hirvonen,~ T h o m a s B. Shows,w a n d Karl Tryggvason*
*Biocenter and Department of Biochemistry, University of Oulu, SF-90570 Oulu, Finland; r Department of Pathology,
University of Helsinki, Helsinki, Finland; w
of Human Genetics, RoswellPark Memorial Institute,
Buffalo, New York 14263; IIInstitute for Biophysics, University of Linz, A-4040 Linz, Austria; and ~Department of
Medical Biochemistry,University of Turku, Turku, Finland
Abstract. We describe the identification of a novel
and domains V, IV, and III are shorter, resulting in a
truncated laminin chain. The alternative sequence had
a shortened domain I/II. In accordance with the current nomenclature, the chain characterized here is
termed B2t. Calculation of possible chain interactions
of laminin chains with the B2t chain domain I/II indicated that the B2t chain can replace the B2 chain in
some laminin molecules. The gene for the laminin B2t
chain (LAMB2T) was localized to chromosome lq25q31 in close proximity to the laminin B2 chain gene.
Northern analysis showed that the B2t chain is expressed in several human fetal tissues but differently
from the laminin B1 and B2 chains. By in situ hybridization expression of the B2t chain was localized to specific epithelial cells in skin, lung, and kidney as opposed
to a general epithelial and endothelial cell expression of
the laminin B2 chain in the same tissues.
1. Abbreviations used in this paper: EHS, Engelbreth-Holm-Swarm;
HSPG, heparan sulfate proteoglycan.
adhesion, and locomotion. The laminin molecule participates in the assembly of basement membranes through binding to other laminin molecules, type IV collagen, nidogen
(entactin), and basement membrane heparan sulfate proteoglycan (2, 69). The cellular activities of laminin are presumably mediated by cell surface receptors, many of which
belong to the integrin family (47).
Identification of new laminin chains, s-laminin, a close
homologue of the B1 chain (34) and merosin, an A chain
homologue (18) has demonstrated that laminin is a considerably more complex protein than previously anticipated. Taking into account the increasing number of laminin subunit
chains, Engel et al. (20) have proposed a new terminology
for the laminin chains. According to this classification, the
classical EHS laminin A chain is termed Ae, merosin Am,
the B1 chain Ble, s-laminin Bls, and the B2 chain B2e. This
terminology is used in this article. Analysis of laminin isolated from human placenta has demonstrated the existence
of isoforms with chain composition Ae-Ble-B2e, Am-BleB2e, Ae-Bls-B2e, and Am-Bls-B2e (21). This is supported
9 The Rockefeller University Press, 0021-9525/92/11/679/15 $2.00
The Journal of Cell Biology, Volume 119, Number 3, November 1992 679-693
679
AMININS are large, basement membrane glycoproteins
consisting of three chains connected by an u-helical
coiled-coil domain. The laminin molecule has a crosslike structure with one long arm and three short arms (69).
Laminin, first isolated from a murine Engelbreth-HolmSwarm (EHS) t tumor (70), was shown to be a heterotrimer
consisting of one heavy A chain of 400 kD and two light
chains, B1 and B2, of ,,o200 kD each (14). The primary
structure of the laminin A, B1, and B2 chains has been determined from mouse (1, 60, 61, 62), man (31, 51, 53, 54), and
Drosophila (12, 26, 48, 49). The laminin chains have a characteristic domain structure with internal repeats (2, 3). The
short arms are formed of EGF-like modules and globular
domains. The long arm is formed by heptad repeats typical
for a-helical coiled-coil proteins. Diverse biological functions attributed to laminin include stimulation of cell growth
and differentiation, and promotion of neurite outgrowth, cell
Downloaded from on June 14, 2017
laminin chain. Overlapping clones were isolated from
a human fibrosarcoma HTI080 cell cDNA library spanning a total of 5,200 bp. A second set of clones contained an alternative 3' end sequence giving a total of
4,316 bp. The longer sequence contained an open reading frame for a 1,193-residue-long polypeptide. The
alternative sequence was shortened at the carboxylterminal end coding for a 1,111-residue-long polypeptide. The amino acid sequence contained 21 amino
acids of a putative signal peptide and 1,172 residues or
alternatively 1,090 residues of a sequence with five
distinct domains homologous to domains I-V in laminin chains. Comparison of the amino acid sequences
showed that the novel laminin chain is homologous to
the laminin B2 chain. However, the structure of the
novel laminin chain isolated here differs significantly
from that of the B2 chain in that it has no domain VI
Published November 1, 1992
human Ble chain residues 1,183-1,785 (53), human B2e chain residues
1,035-1,600 (54), and human Ae chain residues 1,561-2,126 (51). By standard algorithms these regions are predicted to have mainly ~belical structure. In the first step the sequence was ordered in heptad repeats
[a,b,c,d,eJ, g]n so that positions a and d are most frequently occupied by
hydrophobic residues (lie, Phe, Val, Leu, Trp, Met, Ala) and e and g by
charged residues (Arg, Lys, Glu, Asp). As already found for the mouse
laminin chains, several phase shifts have to be introduced to get an optimal
heptad pattern (2). Then the heptad repeats were aligned to adjacem ones
assuming a parallel homodimeric configuration as well as those of human
Ble, B2e, and Ae assuming a heterodimeric assembly. With respect to chain
recognition, the specific distribution of charged residues which can form
salt bridges between the chains appears critical. The number of favorable
and unfavorable pairs of amino acids in adjacent positions were counted as
+1 and -1, respectively. According to the designation of McLachlan and
Stewart (46) the residue pairs were considered 2e-lg', lg-2e', lg-2a', 2a-lg',
ld-le', and lead; where numbers indicate the relative heptad number of
chain n and n'. Their shifts account for the rise of the helix. The interchain
ionic interactions factors were calculated by dividing the sum of interactions
by the number of beptads/chain.
Chromosomal Localization
Human-mouse somatic cell hybrids and procedures used for chromosomally
assigning LAMB2T gene have been described (37, 64, 65). In situ hybridization was accomplished as reported previously (22, 50, 74).
Northern Analysis
Materials and Methods
Poly(A) RNA was isolated from human HTI080 fibrosarcoma cells (ATCC,
CCL 121) and human chorioearcinoma (JAR) cells (24) as described previously (37), ,-05 #g of poly(A)-RNA was run in parallel lanes in a 0.7%
agarose formaldehyde gel and transferred to a nitrocellulose filter (Sleicher
& Schuell, Keene, NH). The filter was cut imo strips which were hybridized
separately with human eDNA probes for the laminin Ae (51), B1e(53),
B2e(54), and the B2t chain identified in this study. The probes were made
by nick translation of the insert DNAs to similar specific activity allowing
approximate comparisons of transcriptional activity.
Total RNA was isolated from 18-19-wk-old human fetal tissues by routine methods (44) with the appropriate approval of the Ethics committee.
Samples containing 10 #g of each RNA were run in a 1.2% agarose formaldehyde gel and transferred to a GeneScreenPhis filter (New England Nuclear, Boston, MA). The filter was hybridized sequentially with the different
human laminin eDNA probes.
cDNA Cloning and Sequencing
In Situ Hybridization
All eDNA libraries used in this study were made from human fibrosarcoma
cell (HTI080) poly(A)-RNA. To obtain clones for the human basement
membrane beparan sulfate prot~glycan (HSPG) core protein, a previously
described specific library (37) was screened with an end-labeled, degenerate oligonucleotide, h22, based on the sequence GQTLDL from the amino
terminus of a short peptide sequence of the human core protein (33). This
resulted in the isolation of a clone, HT2-7, which turned out to code for a
previously unknown laminln-like chain. The HT2-7 was then used to screen
a eDNA library made with both oligo(dT) and random primers. Several
overlapping clones were obtained (Fig. 1) and fragments of these clones
were used to isolate further clones towards the 5' and 3' ends of the eDNA.
To obtain eDNA clones covering the entire 5' end, two primer extension
libraries were made using primers 1322, complementary to bases 199-222
and B23, complementary to bases 177-197, from the 5' end of the L52
eDNA clone. Yet another HTI080 eDNA library made with an oligo(dT)primer was used to obtain eDNA clones for the 3' end.
The nncleotide sequence was determined from both strands, either
manually using dideoxy sequencing (59) with Sequenase (U.S. Biochemical
Corp., Cleveland, OH) or automaticallywith thermocycle sequencing using
Amphq'aq (Perkin-Elmer Cetus Instnmlents, Norwalk, CT) and an automarie DNA sequencer (A.L.E Pharmacia, Uppsala, Sweden). The sequences were analyzed using MicroGenie software (Beckman Instruments,
Inc., Fullerton, CA).
To explore whether the B2t chain might be able to assemble with other human laminin chains, interehain ionic interaction factors were calculated for
the sequence region 613-1,111 with domains II and I of the other chains:
To prepare sense and antisense probes for the laminln B2t and laminin B2e
chains, which would not crosshybridize, fragments from the least homologous 3' end region of the cDNAs were used. For the laminin B2t chain a
PstI-EcoRI fragment of clone L15 (bases 2995-3840, Fig. D and for the
laminin B2e chain a polymerase chain reaction (PCR) fragment with
artificial BamHI-PstI sites (corresponding to bases 105-616 in the 3' noncoding region) (38) were subcloned into pSP64 and pSP65 vectors in sense
and antisense orientation. The 32p-label~ antisense transcripts were
shown to recognize only the specific messages for either the laminin B2t
or the laminin B2e chain in HTI080 cell RNA (data not shown). For in situ
hybridization the antisense and sense probes were labeled with 35S-UTP
(Amersham International, Amersham, UK) using Sp6 RNA polymerase
(Promega Biotec, Madison, WI) for 1 h at 37~ The unlabeled DNA sequences were removed with RNAse free DNAse (Promega Biotec) and
treated with limited alkaline hydrolysis. The unincorporated labeled nucleotides were removed and the probes were precipitated with ethanol, dried,
and dissolved in Wilkinsons hybridization buffer (72) with 100 mM [YVF
to 2 • 106 cpm/ml and used for in sire hybridization.
Human fetal tissues from the 17th gestational week were embedded in
O.C.T. compound-embedding medium (Miles Laboratories Inc., Elkhart,
IN) and 5-~m frozen sections were cut on aminoalkylsilane-pretreated objective slides (56). The sections were airdried and fixed with freshly made
4% paraformaldehyde (PFA) supplemented with 5 mM MgCl2 in 0.1 M
PBS for 15 min at room temperature, dehydrated in alcohol, and alrdried
and stored at -70~ In situ hybridization was performed according to Cox
et al. (15) and Wilkinson and Green (72) with some modifications. Briefly,
the frozen sections were rehydrated in PBS at room temperature for 5 rain,
and treated with 0.5 #g/ml proteinase K (Sigma Chemical Co., St. Louis,
MO) for 7 min at room temperature. The slides were washed with 0.1 M
glycine in PBS for 5 rain at room temperature, postfixed with freshly made
The Journal of Cell Biology, Volume 119, 1992
680
Estimation of Interaction Potential of B2t with Other
Laminin Chains
Downloaded from on June 14, 2017
also by immunostaining of tissue sections which have shown
colocalization of these chains in basement membranes (58).
In vitro de- and renaturation studies with EHS laminin have
shown that the assembly of this laminin is a specific process
and only molecules with one Ae chain, one Ble chain, and
one B2e chain are formed (35, 36). The carboxyl-terminal
parts of laminin chains assemble into a coiled-coil structure.
This structure is stabilized by ionic interactions between the
chains. Calculations of these interactions show that only heterotrimeric molecules with one A-type chain, one Bl-type
chain, and one B2-type chain are favorable (20). Studies on
the expression of laminin subunit genes and distribution of
laminin subunit chains have shown differences in their spatial
expression (4, 13, 19, 39, 40, 51). The existence of multiple
divergently expressed laminin chains suggests different functions for the laminin isoforms.
In the present work we describe the isolation and characterization of full-length eDNA clones for a new member of
the laminin chain family. Despite several unique features,
this polypeptide has considerable sequence similarity with
the B2e chain, although it is substantially shorter. This chain
is therefore termed here B2t (t = truncated). The gene for
the laminin B2t chain (LAMB2T) was localized to chromosome lq25~q31 (25), to the same region where the laminin
B2e chain gene has been previously localized. The expression of the mRNA for the LAMB2Tgene was found to be restricted to only a few human fetal tissues, thus differing
substantially from the almost ubiquitous expression of the
LAMB1 and LAMB2 genes. Furthermore, in situ hybridization demonstrated cell specific expression of the B2t chain
in certain epithelial cells in skin, lung, and kidney, in contrast to a general expression of the B2e chain in epithelial
and endothelial cells in these tissues.
Published November 1, 1992
Lpe47
Lpe45
L52
L4
L7
HT2-7
L71
A30
L 70
L61
As
ATG
B2t-a
I
TGA
I
I
I
H
P
I
P
I
I
H
H
TGA
B2t-b
H
L15
L26
SCALE
(~
10=00
2000
3000
4000
50~)0 lop
Figure 1. Scheme of 12 eDNA clones encoding the laminin B2t chain, eDNA clones with an arrow tail represent clones made by primer
extension. Clone L26 is a mixed clone containing a short region different from the other clones (not shown). Location of the ATG translation
initiation signal and the 3'-end TGA translation stop codons present in the two different set of clones are shown. The 3'-end sequence of
clones L15, L26, and L69 is illustrated by a gray line. Restriction enzyme sites for Pstl (P) and Hindlll (H) are indicated. Scale in base
pairs is shown at the bottom.
4% PFA-5 mM MgC12, and rinsed in 50% deionized formamide (Merck &
Co., Rahway, NJ) and 2 x SSC. The sections were acetylated in fresh 0.25 %
acetic anhydride in 0.1 M triethanolamine for 10 rain at room temperature
and rinsed again with formamide and 2x SSC. The sections were prehybridized in Wilkinsons hybridization buffer containing 100 mM DTT for 2 h
at 50~ The prehybridization mixture was removed and 30--40 #1 of new
hybridization buffer with labeled cRNA probes was added and hybridization
continued at 52~ for 16-18 h. The posthybridization washes were performed according to Wilkinson and Green (72) followed by autoradiography with an NTB-2 film emulsion (Eastman Kodak Co., Rochester, NY).
The sections were exposed under light-safe conditions at 4~ for 7-12 d,
developed in a D-19 developer (Eastman Kodak Co.), counterstained with
Harris' hematoxylin, dehydrated, and mounted with Permount.
The HT2-7 cDNA clone, isolated by screening a HTI080 cell
eDNA library with the degenerate oligonucleotide H22,
contained an insert of ,x,l.5 kb. Initial sequencing of the
clone demonstrated that the derived amino acid sequence has
substantial similarity with sequences of laminin chains. Although the sequence of the oligonucleotide used for initial
screening was based on an amino acid sequence from the human basement membrane HSPG core protein, no significant
sequence homology was found between the amino acid se-
quence derived from clone HT2-7 and the sequence of the
HSPG core protein from which the oligonucleotide was derived. Some homology is found between the sequence coded
by HI2-7 and the laminin-like region of HSPG core protein.
On Northern analysis the cDNA hybridized to at least two
messages of •4.5 and 5 kb. Screening of the oligo(dT)/
random primed library with the HT2-7 insert resulted in the
isolation of overlapping clones L4, L7, and L15. A 5' end
fragment of L4 was used to isolate clone L52 from the same
library. Clone L15 was used to isolate the 3' end clone L26
from the oligo(dT)/random primed library. To obtain the entire 3' end of the cDNA clone L15 was used to isolate clones
L61, L69, LT0, and L71 from an oligo(dT) primed library.
To obtain clones extending to the 5' end of the mRNA,
primer extension libraries were made and screened. This
yielded several clones, the largest being Lpe47, reaching
only 45 bp upstream of the L52 cDNA clone. Primer extension and isolation and S1 nuclease analysis ofgenomic clones
confirmed that this site corresponds to the initiation of transcription (Kallunki, T., unpublished observation). Illustration of the eDNA clones and their partial restriction map is
shown in Fig. 1.
The nucleotide sequence of the overlapping clones and the
predicted amino acid sequence are shown in Fig. 2. As illustrated in Figs. 1 and 2 two types of eDNA clones were obtained for the 3' end region. Clones L61, L70, and L71, con-
Kallunki et al. Novel Laminin B Chain
681
Results
Cloning of cDNA Encoding a Distinct
Laminin-like Chain
Downloaded from on June 14, 2017
A2o
L69
Published November 1, 1992
a
1
1
268
51
G
M ~
F
A
L
W L
A
G ~ L ~ ) P
S
~
L
~
L
L
C
P
C
A
A
C
"
A~T
CTGCAATGACAACACTGAT(~CATTCA~~TGG~r
N ~ N
D M T D G I H ~ E
K ~ K
N
R ~ L
418
~c~-,~c~c~cc~c.c~,~,~c.~a~cc.~_a~ccacc~
718
201
(~%ATACAGTGTCCATAAGATCA~AC ~ T C A K G A T G T T G A ~ C
g Y 8 V H K I T S T F H g D V D G W K
C
S
T
R
G
S
~;~-*-~.GF
Y R
E
C
A
G
C
VI~D~)N
G
DOMAIN V
H R E
a
D
G G A G A C a n A G A L ~
K
8
R
~{~l
F
D R
E
L
H
R
Q
T
e
117
N
T T G T ~ A G T G C T C ~ A T G T C . % C A ~
P ~ N ~ N
S K G ~ L ~ A R ~ D
N $
R ~ L
c~c~_.~~.~cc~c~.~c.~.~c,~c~.~o,~cc.~u~-~c,~~'~c~
a
50
~
417
i00
56~
.
DOMAIN IV
868
251
A
K
1018
301
ATGCCACTT~
M P L G
K
T
1168
351
ACATATGC.AGAATACAGTACTGGGTACATTC,
T Y G E u S T G Y I
1318
A A C ~ T T C A ~ C T ~
146~
451
~e~cc~ccCC~GCC~rercc~r
N D P H D P R S ~
161~
~-~c~cccc-n~u~c~v.cccc~u~cc~~c.~c~Gc~&sccc=~c~-~a*n~-~
r u 0 ~ r = ~ a u v v R ~ 9 0 ~ 9
9
1768
A
L
(~CCA
551
1918
601
~
T~TCAAC~GGT~ATGGGCAAAGCCTGTCCTTT
F L G N Q Q V S Y G Q S
P
501
F
~
G
A
9
M
~
G
~
A
r
~
G o
ACAATGT~
D ~
L
~
S
A
R
P @
P ~
H
N
P
V
.
G
S ~
F
CCAAATGGCTTTAAAAGT~GC.AGGC
P N G F K S L A Q E
,
E
G
R
S
G
V
R
__
S
G
A
~
~%~ACTA
D Y R
V
S
G
D
$
S
A
~
.~rc~
V I
N
V
A
~
A
D ~
9
K
L
R
G
G
R
P
Q
L
D
P
V
Y
867
250
R
I
T
A
P
L
1017
300
L
S
ACTTT~ATCC~ACTGC~TCTCACAGCCC~TCCGAGCT
Y F E u R R L L R ~
A L
R
I
R
A
1167
350
.CCCT~CAGTGTATAT~ACAAGGGGCAATTC~TTGT~AC
P W V E Q ~
I 9 P V G Y K G Q ~ 9
DOMAIN Ill
TCCAGACAC~TTGTTATT~TC,
AGAATCCTGACA
.
P
A
Plg
D
R
.CACCC.~TCTGCCCATGATG'TC,~~~TCA~
M P S A H D V I L E ~ A G
.
N ~
A
0
A 9
M
I
D
8
g
CACAAGATTA~CGTTGA~TCA~AACAT~
T R 5 A E S M V E S
AGCCCC,
GACGGTG~GCAA
S P D G A v V Q
A
~
T
E
E
V
V ~
S
A
8
=
.
r~
N N ~
9
~
P
P
Z
9 G
u
9
cce~r~
T G A
G V
Q
R ~
T'~C~
L E
9
M
~
9
~
~
L
~
0
S
R
z
M
v
P
E
R
E
s
o
G ~ 9
L ~
S
S
E
A
S
L
G
N
.CTAACATTCCTGCCTCAGACCACTACGT~
T N I P A S D M Y V G
CTC~CTA
T E D
Y
$
K
Q
A
L
R
K
T
Q
A
E
T T ~ T A C ~ A
I E A D R S
u
T
R
M
M
Q
L
A
I
T
Q
M
Q
L
$
L
A
E
A
S
N
M
E
Q
L
T
R
E
K
T
K
s
L
_ CCAG~CAAGGC~
A Q O L T R E
~
A
$
L
E
E
A
K
R
TCAAACA~TTCAC~G~AACCA~TAT~GATC,
I K Q K A D S L S S L V
CAGCAGCTCTTACAGAATGGA~GT~GAGAC~TCAGA
Q Q L L Q N G K S G R E K
S
D
Q
L
L
$
R
A
_ TCTTGCTA~AAGCAC4%GCACA~GCACT~ATC~TGCCAL"
N L A K S R A Q E A L S M G ~
E
E
A
M
K
R
L
$
Y
AGATTC~ ~
E I E Q
E
%~ ~0 C~%G ~~fI ' ~ C T T C ~ T G T G A
I G S L N L E A ~
R
V
L
D
AA
N
W
K
E
E
A
2968
951
I
L
K
N
L
R
E
F
D
3118
1001
GL~TGCACAGAGC~T
A A D A Q R
A
K
N
326Q
1051
A~TGAC.A
S E M
E
G
3418
1101
ACATTAC.ACC~CT~TCTC,
ATGC,A C C A G C C T C T C A G T G T ~ T G A A ~ A G G G G ~ A
T L D G L L H L M D Q P L S V D E E G L
3568
1151
C~GCAGA~CCTCCATTTGCT~CAA~TAC~ATGGGATTCT~T~C~AACTT~TTAGGGACAACCTGCCCCCAGGCTGCTACAATACCC~~~T~TA~
Q Q R G H L H L L E T S I D G I L A D V K N L E N I R D N L P P G ~
3718
TCAACTGAGGTTCTTGGGATACAGATCTCAGGGCTCGGGAGCCATGTCA
3868
T T G C A C C A T A C T C ~ T G C T G G G C A T C ~ A G G C A G A T A G G C A ~ ~ T C A A G G A ~ C ~ C ~ % T A ~ A % C T G G A T G G A A A ~ ~ ~ T A ~ ~
4018
CCTG~TTTGGACAAGTGCTGTTGGGATATAGTCAACTTATTCTT~AAT~TGACTAAAG~CT~
R
E
V
R
K
A
E
A
_GAGCTGGAAAGGAA
E L E R K
E
L
E
F
D
~-~zTI aTC,AAGTTGAC~AGC
F Y E V E S
3117
I000
R
A
L
~
S
A
CA ~CAGA
A D
G
A
L
A
T
M
C ~
E K
G
L
A
8
L
K
3267
1050
CA~GAATAT~T~ACAGAT~TTACAC~AAGCCCA~AAGGTTC~ATACCAGAGCCAAGAACG~A~TCCAAGACACACTCAA~
T N M D A V Q M V I T E A Q K V D T R A K N A G V T I Q
D
T
L
N
3417
1100
R
3567
1150
S
R
A
CCCAC~TCAACAGC
T Q I N S Q
K
u
N
T
G
L
R
P
TGATGTCAGAGCTG~GAC~CGT
M M S E L E E R A
Q
A
L
E
Q
TGGG~TTTGAACATGTTTAAT~ATGCTCA~CTGACCTGACCCCATTCCTC,
Q
3717
1193
*
ATCCCATGGCCAG~A
3867
4017
TC,AAATTCTTCCTAA T G T C A ~ C A C ~ C C C A G T C A C A C T G T C ~ C A G T A A A A
TA
4167
4317
4168
4318
286"/
950
E
5
A
2817
900
G
A
K
D
2667
850
Q
Q
S
~CGC
R
Q
E
V
L
T
L
K
8
2517
800
K
L
Q
H
AGTTCA~ACACAAAAGAATCT~
E F K R T Q K N L
D
2367
750
D
V
S
Q
2217
700
$
GGGGC C C ~ / ? ~ C C C ~ T C T C C A G T C ,
G A G E A L E I S $
X
'rGACCTCAAC.ATG
D L K M
GGCCCTGC~TGAAGGA
A L H E G
2818
901
N
2067
650
Q
L
v
D
600
A ~
E L E
Y
G
Q
V
1917
~
u
V
L
G
T
T
176~
~50
~ 9
K
G
Q
F
Q
1617
500
G
A
S
L
r~rAc
D G Y
A
D
K
CTCCTGC, A T T C A ~
L L D S V S
Q
1467
G
L
2668
851
V
TTGGTTTCTAC
$
A
K
G
1317
400
Y
.~GAAC.AGCTA
R S Q E N S
AGCAC,A
A S R S
E
Q
G
~
F
v
L
S
.GCCAGG,A
~
~ a r ~
9
L
P
A
AGTACCTC,A
V V P O
Q
.
D ~
.uc~-n-~rC:rA~CC~C~c~c~cc~cr~c~c
R 9 ~ ~ 9 ~ a . 9 A G ~
G
.GTC.A
S D
L
GA~CTGTAACCCCAT~GCCTGTAGGAT~&~GATGGCA
~
GGA~GTACCAGAACCGAG~TACTCAC~TCACTCAGATG~
G S Q Y Q N R V R D T H R L
L
P
TC~TCA~TCAAC.AT&TGTTT~CTAGA~AT
Q W S Q R H Q D V F S ~ A Q
~
C
A
C
C
~
G
-
f
T C ~ C C , ! ~ T C ~ T C ~
AA'~T~T~CTT
AC~TTGCAT~ATT
A
~
A
~
~
A
~
A
'A~o (CIO,O
C
~
;
~=[ % ~
4467
L71)
4468
T T T C A A A G T G A T A G A A A A G T ~ T T C ~ G A G G T A A A A T T C T C T A C ~ A T T T A T T A G T C C T A A T T C A A ~ ~ ~ T ~ T ~ A ~ A ~ ~
4617
4618
T C C A C C C A T A A T A ~ T ~ A C T C A C A C T ~ G C T ~ C A T C C A T C C C ~ T T C A ~ ~ ~ A C ~ ~ T A T A ~ A ~ A C ~ A ~ ~
4767
4768
TGGGACAGTGGTGACAT
4917
5067
4918
5068
A~~GGAACCAC~AC.C.C--%CTTC
C A C C ~ ~ C T A T G ~
ATTTCCITGGAT~GTr21TTT~AAAT
AAAC.AACAATTGTTAGATCC
CAAAAA
5200
b
~
G
3446
1110
~
M
'
*
~
J
~
T
A
T
A
C
C
T
A
3568
TC~GGCCCAGATAA
3718
ACAC.AACACC.AGAC-AGC~TCCTGCTAAAC~GGACC
3868
GGGTTAD,A G A G A A G G C A G G T T G A C ~ A C T A C T T A A G A T A T T G T T ~ T A A T T G A A G A ~ T C ,
A
4018
~
C
G
C
C
C
C
G
C
A
A
A
A~GGATATCAGTAAATG
ACATTTACTGGACCCI~; L + ~ - ~ - L ~ C C G T T G C T A A G A T ~ C A C C T A T T G C A C T T ~
CAGGGTGT~I~CAOQCCTGTAATTCT
lu R l p e a l
A
G
CA~x-FF~mAG~TAACATCACCACTGT
C
A
~
T
C
A
~
Volume
AAAC,G T ~ C A A A G A A C A G G T G T A T A T
C
~
C
A
T
G
~
TGTGAAGC, A A G A ' F I ~
C
C
~
C
T
~
GG
"
ACT AAAAGT A C A ~
.GTCA~C~TC~
~ A ~
3567
1111
3717
ATATTT C A G A A A C . ~ T C A C T A G ~
AAG~AG~A
~
~
3867
4017
4167
43t6
4168
The Journal of Cell Biology,
A
119,
1992
682
Downloaded from on June 14, 2017
2368
751
C
~
A 9165 N 0 V K ~
DOMAIN t/l|
. Q C . . A C A ~ ~
A L g D I L R D A 0
~
V
~
SI 9
2218
701
V
F
R
CCAAGACTTACACATTC~GGTTAAAT~TCCA~TAA
T K T Y T F R L N E M P S
L
K
GGCAGGATGCAGCAGGCTGA
G R M Q Q A E Q
2518
801
$
~
Q
~-x-~-~C-GC~ATTCCTTGT
2068
651
T
A
~
A C ' I ~ T T G ~ C C C A
~ 0 9
~
P
L
A
A
Published November 1, 1992
Domain Structure of a Truncated Laminin-like Chain
The laminin-like chain identified here has domains similar
to those found in laminin chains and the domains are numbered accordingly (Fig. 3). Domain V (residues 28-196) is
IVb
IIIb
l,,vvv,
IVa
j~.~{"~/~
NH2 c~ ~ N H
i
(
)
(
1/11
c
(
c
c
c
Ble
Chain
)
c
B2e C h a i n
c I COOH
Ae
c
B2t C h a i n
c
COOH
c
Chain
S e q u e n c e identity of
Laminin B2t d o m a i n s
H u m a n B2e
vs
III IV V
(
(
(
(
c
3
2
V
55
IV
52
III
55
1/11
34
Human Ble
36
17
40
27
H u m a n Ae
37
28
41
22
Figure 3. Structural domains of the laminin B2t chain and comparison with the laminin Ae, Ble, and B2e chains. A schematic model
of the structural domains in laminin chains is shown at the top. At
the bottom comparison of laminin B2t chain structural domains
with similar domains in the laminin Ae, Ble, and B2e chains. Percentage of identical residues in the aligned sequences between
the domains is shown inside the box representing the structural
domain.
formed of three and one half cysteine-rich EGF modules
characteristic for laminin. Domain IV (residues 197-381) is
a 190-amino acid region containing a single cysteine. This
domain is likely to form a globular structure similar to
domain IV in laminin. Domain III (residues 382-608) is
formed of the second half of an EGF module and four additional modules. Domain I/II of the longer polypeptide
(residues 609-1,193) starts w i ~ two closely spaced cysteines
and contains one cysteine close to the carboxyl terminus. It
is an a-helical domain of 585 amino acids with a heptad repeat structure typical for coiled-coil proteins. In the shorter
polypeptide form domain I/II is shortened at the carboxyl
terminus by 82 residues.
Alignment of the sequences of the structural domains in
the laminin-like chain with similar domains in the laminin
Ae, Ble, and B2e chains reveals that there is high sequence
Figure 2. Nucleotide sequence of the laminin B2t chain cDNA and the derived amino acid sequence. First line, nucleotide sequence of
the cDNA clones. Second line, deduced amino acid sequence. (A) The putative signal peptidase cleavage site; (bent arrows) borders of
structural domains, cysteine residues are circled, and potential attachment sites (Asn-X-Ser/Thr) for oligosaccharides are boxed. (a) Nucleotide and derived amino acid sequence of cDNA clones encoding for the 5,200-bp sequence. (b) Nucleotide and derived-amino acid sequence
of the alternative 3' end sequence from cDNA clones providing the sequence for the total of 4,316 bp. These sequence data are available
from EMB/GenBank/DDBJ under accession numbers Z15008 (a) and Z15009 (b).
Kallunki et al. Novel Laminin B Chain
683
Downloaded from on June 14, 2017
tained a sequence (Fig. 2 a) which gives a total length of
5,200 bp. Clones L15, L26, and L69, contain a sequence
(Fig. 2 b) which differs at the 3' end giving a total length of
4,316 bp. The first sequence (Fig. 2 a) has a ll7-bp 5' untranslated region, an open reading frame starting from position 118 with the first ATG codon for methionine and ending
with a translation stop codon TGA at position 3,697 and a
1,504-bp 3' untranslated region. The 3' untranslated region
contains two conventional polyadenylation signals AAUAAA at positions 4,841 and 5,172. The sequence ends 18
nucleotides after the second signal with a possible poly(A)tail of only five nucleotides. One of the clones, L71 ends with
a poly(A)-tail of 30 nucleotides which begins 15 nucleotides
after a slightly unusual polyadenylation signal AUUAAA at
position 4,413. The two clones with different polyadenylation sites give mRNAs of 4,463 and 5,200 bases.
The second and shorter sequence (Fig. 2 b) differs in the
3' end starting from position 3,446. It gives an open reading
frame ending with a translation termination codon TGA
at 3,451 and 866 bp of 3' untranslated region giving the
total length of 4,316 bp. Computer homology search revealed that the last 300 nucleotides contain an Alu repeat
with a poly(A)-tail at the 3' end.
The 117-nucleotide region preceding the ATG codon has
an open reading frame, but we surmise that this ATG codon represents the true translation initiation site for the following reasons. First, extensive screening of primer-extension cDNA libraries did not reveal clones extending further
upstream from the Lpe47 cDNA. Second, the sequence surrounding the ATG agrees well with the consensus sequence
for the vertebrate translation initiation cgccA/GcCAUGg
(10) although it has a C following the ATG. Third, this initiation site is also supported by analysis of the amino acid sequence following the methionine, which shows a stretch of
hydrophobic amino acids typical for a signal peptide. Computer program analysis predicting the signal peptide cleavage
site according to von Hejne (70 b) suggested a cleavage site
after Ala21.
The open reading frame of the first and longer sequence
(Fig. 2 a) provides for a 1,193-amino acid polypeptide. The
open reading frame of the second sequence (Fig. 2 b) provides for 1,111 amino acid residues. After cleavage of the signal peptide the mature polypeptide chain would contain either 1,172 or 1,090 amino acids with a predicted molecular
weight of 128,650 or 119,280, respectively, for the longer and
shorter sequence. There are six putative Asn-linked glycosylation sites Asn-X-Ser/Thr (Fig. 2). If all of them are used,
the molecular/mass could be closer to 140 or 130 kD.
Published November 1, 1992
ngt
28
B2e
342
B2t
88
B2e
402
B2t
148
B2e
454
DE 9
ngt
206
nge
514
KITSTFHQDVDGWKAVQR~--~PAK
I
III
I
III
SISSTFQIDEDGWRAEQRDGSEAS
B2t
266
B2e
574
B2t
326
B2e
634
Dmt
386
B2e
694
B2t
445
rage 754
B2t
503
B2e
814
i9169
Q E e Y FI :
RS
I
GP 9
VGYKGQF 9169
II I
I I I I:
I
:II
I
AD
I
LESSN
ALI
I I I
SKAQGGD
B2I 1054
8 L I ANLGT
mmt
QSRLDD
I I I
RG 9169169
I
SV 9
Y
L Q W S Q R H Q D V F S S A Q R L D P V Y F V A P A K F L G N Q Q V S Y
I : I I
I I :
"
I
I I : I I I I I I I
I
L E W S S E R ~ D I A V I S D S Y F P R Y F I A P A K F L G K Q V L S Y
:1
E G A G L R I T A P L M P L G K T L P 9
I I I I II:
I I
I
I
I
I
I I I
I
E G A G L R V S V P L I A Q G N S Y P S E T T V K Y V F R L H E A T D Y
I I
I
I
I I
I : I I I
I I I I
GTYSERBAGYLDDVTLASARPGPGVPATWVESI 9
II
I
F G T 9 1 6 9 1 6 9 1 6 9 1 6 9 1 6 9
I:
I
I:I
I I I:I
I I I :
I:
I
I I
G S S9
IIII
I I I
I
I I I I I I
K 9169169
I :I
I
I
I
I
IV
I : I
KT
I
KE
I I I I
VV 9
I
I I I
I I
9
I
I:
I
I
I I I
I
I I
I I I I I
N9
T ~ T T ~169
L9
I I I I I
a Yr G
I I I : I I I I
I I I I I
I I I I
I I I
I I
GQD@GA~D
P~
u N LQ
OF
E G~KP~D~HP
III
I : I I : I I
I
Y GTMKQQ
S S9
VT
GQ 9169
HVT
P e 9G G P
I I
I
N9
LKMTVERVRALG
R~E
H -- -- G A F
VNHF
S~P
GP
A~YNQVK
I QMD
8 GQ
G9
E O S L S L
QF
MQQ
L?RME
I :
GVVP
D TE LE GRMQQ
I I
I
I
I
I :
GDEMVTDQAFEDRLKE
I I
I
A S G N 9 1 6 9 1 6 9 1 6 9 1 6 9
S D G T 9169
I I
I
I
I
I I I
I
P G I T GQH~E
_9
V
:
A , Q A L Q D I L ~ D ~ Q ~ S Z O ~ S R S L G L Q L ~ V ~ S ~ , . S ~
I I
I
I I : I I
I
I
I
AE R E VMD
L LR E AQ D VK D VD Q N LMD
R L Q R V~N-~L
VRDTHRLI
SQYQNR
TQMQL
S LAE
SEAS
LGNTN
I
I
S 6 Q
P ASDHYV
B2e 1114
DDt
750
B2e 1172
B2t
810
NMEQLTRETEDYSKQALSLVRKALHEGVG
GPNGFKSLAQEATRLAE
SHVE
SAS
:
: I
I
: I
I
I
I I
I I : I I
I I I
I
"
I
D p N~--M-~L L A E E A a K L A E a H K Q E A D D i V R VA K T A~-D--~S T E A Y N L L L R T L
GAVVQG
LVEKLEKTK
ATQAE
S LAQQLTRE
I
I :
9"
ngt
867
.,,.,
"
ARVHE
B2e 1226
QVEEAKR
IKQKAD
SL
IEADRSYQHSLRLLDSV
AK
9
B2t
925
AN
L A K S R AQ
IDA
B2t
SDKTQQAERALG
rome 1402
E A L S M G~A--~F
Y EVE
A
NEKI
TI R E A l
B2t 1047
AS
B2e 1462
NNMLKQLQEAE
B2t 1107
HLMDQP
B2e 1583
IIII
:
LD
SETLE
.
.
: :
I
S I L K N L R E F D L Q V D N R K A E A E E A M K R L S Y I S Q K V S D A
9
"
1 1 : : I
~N~I
I : : 1
AEEALRKIPAI~-Q-~ITEA
[HE
QQAL
I
I I
KE
I I :
LKRKQDDADQD
LSVDEEGLVLLEQKLSRA
B2e 1522
ILAD
I
IMKD
IYASVAQLSP
,,,
AGEALEISSEIEQEIGSLNLEA~---V-~ADGALAMEKGL
SAAADAQRAKNG
LKSEMREVEGELERKELEFDTN
:
B2t 1165
,,
LQGVSDQSF
DYEDLREDMRGKELEVKN
B2e 1342
987
AGDKAVE
A G E~-~
SP
E F K R T Q K N L G N W K E E A Q Q L L Q N G K S G R E K S D Q L L S R
S SLVTRHMD
N
B2e 1284
9
8GSGSPD
VKNLE
N IRDN
I
I I I
I I
IRNLE
D IRKTLP
LPP
G9
I I
I I
S G9
:
I
M D A V Q M V I T E A Q K V D T R A K N A G V T I Q D T L N T L D G L L
I
II
:
I
I
:
I
: I
M M M A G M A S Q A A Q E A E I N A R K A K N S V T S L L S I I N D L L
SQLRPMMSELEERARQQRGHLHLLETSIDG
l
I
l
I I
l
l
D E M K V S D L D R K V S D L E N E A K K Q E A A I M D Y N R D I E E
:
II
KTQIN
l
LEQQ
I I
I
N T P 8 IEKP
Figure 4.
A l i g n m e n t o f the h u m a n l a m i n i n B2t a m i n o acid s e q u e n c e w i t h the h u m a n l a m i n i n B 2 e (54) a m i n o acid sequence. T h e s t r u c t u r a l
d o m a i n s are boxed and indicated by R o m a n n u m e r a l s o n the right. T h e a m i n o acid s e q u e n c e of b o t h c h a i n s is n u m b e r e d f r o m the initiator
m e t h i o n i n e . All cysteines are circled a n d the g l y c o s y l a t i o n sites N-X-T/S are boxed.
The Journal of Cell Biology, Volume 119, 1992
684
Downloaded from on June 14, 2017
934
Np
DGAH 9169169169169
A T Y G E Y S T G Y ~ D ~ - ~ L I S A R P V S G A P A P W V E Q ~
I
I
DPFGEHGPVRP 9169
II
I
I I I I
DPLGRNGPVRL 9169
B2e
692
I
9 1 6 9 1 6 9 1 6 9
I
I:I
I
I I I I I I I I
632
I
9169169169
APNPADK 9169169
N
Y ~ N L D G G N P E G 9 1 6 9 1 6 9
~W-~PQLSYFEYRRLLR~--L~ALRIR
I
I
I
II:
II
PWRPALTPFEFQKLLN~-L-~SIKIR
Ap
994
N9
R 9 1 6 9 1 6 9 1 6 9
I I
~ 1
I I :
GQSLSFDYRVDRGGRHPSAHDVIL
II
III
: l l l l
GQ~-~FSFRVDRRDTRLSAEDLVL
874
B2t
l
FF
563
B2e
I
I I~,~176
DAGR 9169169169
B2t
585
D P E L YI IR I
SAR 9169169169
SKG~L
B2e
B2t
N 9 1 6 9 1 6 9 1 6 9 1 6 9 1 6 9
G KSRQ 9169
9169
IIII
Published November 1, 1992
Table L lnterchain Ionic Interactionsfor a Parallel,
In-Register Chain Arrangement of B2t Domains with Those
of Other Human Laminin Chains
Chain assembly
B2t-B2t
B2t-B2e
B2t-Ble
B2t-Ae
Ble-Ble
B2e-B2e
Ae-Ae
Interchain interionicinteraction
factor
-0.152
+0.013
+0.207
+0.198
-0.105
+0.000
- 0.116
values are -0.105, 0.000, and -0.116, respectively. In contrast, the values for the association of B2t-Ble and B2t-Ae
give positive values of over +0.1. Values of similar magnitude have been calculated for the long arm COOH-terminal
fragment E8 of mouse EHS tumor laminin (20). In vitro disand reassembly studies performed with the fragment (35, 36)
are in complete agreement with the predictions based on the
calculations. The low positive value for B2t-B2e interaction
suggests this as an unprobable chain arrangement. Accordingly, it seems likely that the B2t chain could replace B2e
under certain conditions. The values calculated for the other
laminin variant chains, Bls and Am chains, also suggest that
they replace their counterparts Ble and Ae, respectively (20).
Chromosomal Localization of the Laminin B2t Chain
Gene (LAMB2T)
The gene for the laminin B2t chain (LAMB2T) was mapped
to chromosome 1 by using a cell hybrid panel of 35 somatic
cell hybrids. The hybridization of the partial cDNA clone for
LAMB2T correlated with the presence or absence of human
chromosome 1 (data not shown). Examination of cell
hybrids that retained regions of chromosome 1 localized the
gene to lql2~qter. In situ hybridization of the cDNA to
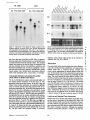
metaphase chromosomes confirmed the localization to chromosome 1 (Fig. 5). The majority of signals localized over
bands lq25-'*q31.
Northern Analyses
Tentative Interaction of the Laminin B2t Chain with
Other Laminin Chains
To estimate whether the B2t chain can assemble with itself
or other known human laminin chains, we calculated the
interchain ionic interaction values for different parallel inregister arrangements (Table I). As found for all other laminin chains studied so far, the negative value of a homodimeric
associate indicates this as a rather unfavorable configuration.
For human Ble-Ble, B2e-B2e, and Ae-Ae the corresponding
Expression of the laminin Ae, Ble, B2e, and B2t chains was
compared in cultured cells and human fetal tissues by Northern analysis. The expression pattern of the different laminin
chains in the HTI080 fibrosarcoma cells and human choriocarcinoma, JAR, cells is shown in Fig. 6. In the HTI080
cells, the laminin Ae chain is expressed only at a low level.
Hybridization with the Ble, B2e, and B2t chain cDNAs
yielded signals of about equal intensity. The B2t cDNA
probe revealed two distinct bands of ~4.5 and 5 kb. The JAR
cells showed strong signals with the Ae and Ble cDNAs and
slightly weaker signal with the B2e cDNA, but no signal
with the B2t cDNA. Comparison of B chain expression in
a number of human fetal tissues is shown in Fig. 7. The Ble
and B2e mRNAs were generally expressed coordinately in
most tissues, except in the brain where the ependymal and
intermediate zones and cortical plate displayed Ble chain signals much weaker than the B2e chain signal. Expression of
the B2t chain mRNA in these three tissues was negligible or
absent. In skin the expression of the Ble chain was consider-
Kallunki et al. NovelLamininB Chain
685
Downloaded from on June 14, 2017
identity with the laminin B2e chain. Domains V, IV, and III
are >50% identical between the two chains (Fig. 3). The
laminin Ble chain is much less similar, with domain IV
showing little similarity. Although the amino acid sequence
of the laminin-like chain characterized here demonstrated
extensive similarities with the laminin B2e chain, it is considerably shorter. We therefore term this truncated laminin
B2e-like chain B2t.
Comparison of the amino acid sequence of the B2t chain
with that of the B2e (54) chain shows (Fig. 4) that the B2t
chain has several features which clearly distinguish it from
the B2e chain. A major feature is that the B2t chain lacks
an amino-terminal globular domain VI present in the other
B chains which forms the globular structure at the end of the
short arms. Another distinct feature is that domains IV, III,
and I/II are shorter. The 21-amino acid signal peptide in the
B2t chain is followed by domain V. The three EGF-modules
can be aligned with modules 2-4 in domain V of the laminin
B2e chain, with the exception of an insertion of eight amino
acids after the second module in laminin B2t chain. The
fourth EGF module is interrupted by a cysteine-free region
of ~190 amino acids, domain IV, similarly to the corresponding domain in the laminin Ae and B2e chains. This is
followed by domain III, corresponding to domain III in the
other laminin chains, containing three complete EGF modules, which can be aligned with modules 7-9 in domain HI
of the laminin B2e chain (54). The fourth module in the B2t
chain contains only six cysteines and it cannot be totally
aligned with module 10 in the B2e chain. The laminin B2e
chain contains two additional EGF modules in this domain.
Similarly to the other laminin chains, the carboxyl-terminal
end of the protein, domain I/II, begins with two closely
spaced cysteines, which participate in interchain disulfide
bonds in the center of the cross in the laminin molecule.
Also, as in the other laminin chains, the amino acid sequence
of domain I/II can be written into heptad repeats, a structure
typical for coiled-coil proteins. However, similarly to the
B2e chain the B2t chain has no domain c~ which is present
in the Ble and Bls chains. The sequence similarity between
the B2t and B2e in this domain is lower than that between
the other domains. In the longer form B2t chain this domain is 585 residues as compared with 579 residues in the
B2e chain. Domain I/II in the shorter form of the B2t chain
is 503 residues and has no carboxyl-terminal cysteine residue, which has been shown to participate in the formation
of a disulfide bridge between the two B chains in the EHS
laminin (52).
There are only six possible glycosylation sites in the B2t
chain as opposed to 14 in the B2e chain. In general, the
glycosylation sites in laminin chains are concentrated to domain I/II. In the B2e chain there are nine sites in this domain
but only two in the B2t chain.
Published November 1, 1992
a
30
20
10
T P~ .q
~
r
i; l. q.m p .Iq.. P ~ : [ p [.),1;
1,LTI;L 1
0
E
Z
3O
20
I0
I~ . dll . m..
10
it
9
.
.m
L
Jl.
...m
l.l~ l.lq ]~ i~ ~ I*,l" ,i;.. Pr"
"i~,9
l;z[o~IU.*IU~I
2
.
Chromosomes
Downloaded from on June 14, 2017
b
gO
P
O0
IO
q
N
I
Figure 5. In situ hybridization of the HT2-7 cDNA to metaphase
chromosomes. The histogram (a) displays data for all signals
counted in 50 metaphases. The idiogram of chromosome 1 (b)
shows the distribution of signals on that chromosome and the assignmerit of LAMB2T gene to lq25-'31.
Published November 1, 1992
ably lower than that of the B2e and B2t chains. In general,
the tissue expression of the B2t chain was considerably more
restricted than that of the Ble and B2e chains. The strongest
signals were observed in skin and lung, but expression was
also seen in kidney, thymus, choroid plexus, cerebellum, and
the brain intermediate zone. In contrast, negligible or no signals were seen in the testis, pancreas, adrenal tissue, cardiac
muscle, spleen, liver, calvarial bone, neuroretina, olfactory
bulbs, brain ependymal zone, cortical plate, or meninges.
Cell-specific Expression o f Laminin B2t and B2e
Chain m R N A s in Human Fetal 1issues
Figure 7. Expression of laminin chains in human fetal tissues. Sampies of 10 #g of total RNA from normal human fetal tissues isolated
at ,o18-19 gw were run on a 1.2% gel and transferred to a GeneScreenPlus filter. The same filter was hybridized with cDNAs for
different laminin chains as described in Materials and Methods.
nephrons at the S-shape stage and also in the vascular endothelial cells (Fig. 10, A and B).
Discussion
Five genetically distinct laminin chains have been characterized so far. All the previously identified A- or B-type laminin
subunit chains have a conserved domain structure and
closely similar organization of internal repeats within each
type. In the present work we have characterized a previously
unidentified laminin chain, extensively resembling the B2e
chain. The novel laminin B2t chain differs, however, from
the B2e and other B-type chains by having a truncated structure and, consequently, a considerably lower molecular mass
than the other B chains (130 vs 190-200 kD). The smaller
size of the B2t chain is due to the absence of a domain corresponding to domain VI of the other B chains, but also due
to considerably shorter domains V, IV, and III. As such,
these differences suggest a distinct biological role for the B2t
chain. Additionally the present data indicate that the B2t
chain exists in two different forms, the smaller form having
a shorter domain I/II giving a molecular mass of 120 kD.
The in situ hybridization analyses demonstrated highly cell
and region specific expression of the B2t chain in 17thgestational-week human fetal tissues. In the skin, lung, and
kidney tissues studies, expression was confined to epithelial
cells. In contrast, the B2e chain was expressed both in epithelial and endothelial cells. In skin B2t chain expression was
observed in the entire epithelium with particularly strong
signals in the appendices and adnexes which form the glands
and ducts (Fig. 8, d and e). The B2e chain was expressed in
the epithelium but also in vascular endothelial cells and possibly also in dermal cells (Fig. 8, A and B). In lung the B2t
chain was expressed exclusively in epithelial cells of bronchi
and alveoli (Fig. 9, C and D). The B2e chain was highly expressed in alveoli and also in vascular endothelial cells, but
only a faint signal was seen in the bronchial epithelium (Fig.
9, A and B). In kidney the B2t chain was expressed at low
level in the collecting tubules in the medulla (Fig. 10, C and
D). The B2e chain was expressed mainly in secretory
Long A r m and Potential Chain Interactions
To date, there are no protein data available on the B2t chain,
and the mode of association of the laminin B2t chain with
the other laminin chains could not be determined in the present study. However, the interchain ionic interactions calculations indicated that the B2t chain can replace the homologous
B2e chain in laminin molecules. Although the sequence
similarity between B2t and B2e is weaker for domain II/I
Kallunki et al. NovelLamininB Chain
687
Downloaded from on June 14, 2017
Figure 6. Expression of the laminin B chains in cultured cells.
Northern analysis of poly(A) RNA from HT1080 fibrosarcoma
cells and human choriocarcinoma, JAR, cells. 5 #g of poly(A)enriched RNA was electrophoresed on 0.7% agarose gels and transferred to a nitrocellulose and hybridized with cDNAs for different
laminin chains as described in Materials and Methods.
Published November 1, 1992
Downloaded from on June 14, 2017
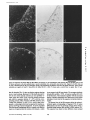
Figure 8. Expression of laminin B2e and B2t mRNAs in fetal skin. In situ hybridization with antisense RNA for the B2e chain (A and
B) shows expression in the skin epithelial layer, vascular endothelial cells, and possibly also dermal cells. Signals for the B2t chain can
be seen in keratinocytes but they are particularly strong in the appendices and adnexes which form the glands and ducts, while vascular
endothelia are negative (D and E). Sense RNAs for either B2e (C) or B2t (F) chains used as control show no signals. Bar, 110 tim.
than for domains III to V, there are distinct regions showing
more or less homology. Residues 613-766 which directly follow the two closely spaced cysteines are most probably located within the center of the cross-like laminin structure
and exhibit a sequence identity of ~ 37 % with the corresponding portion of B2e. Residues 794-908, however, have
a rather low similarity, or only ,~15%. Such a value corresponds to a homology which can be expected for random sequences showing a heptad arrangement. It can be concluded
from the heptad arrangement that in a complex with Ble this
region is arranged around the cysteine-rich domain cr which
is not present in the B2-type chains. The strongest similarity
between B2t and B2e, ~ 5 5 % , is found at residues 921-1,011
of the B2t chain. This similarity together with the increased
density of putative interchain ionic interactions in this region
indicates a crucial role for this sequence in laminin chain assembly.
The shorter form of the B2t sequence lacks the carboxylterminal region corresponding to fragment 25 K of mouse
EHS laminin and also the terminal cysteine which forms a
disulfide bridge between B2e and Ble in EHS laminin (52).
This sequence was found in three different clones isolated
The Journal of Cell Biology, Volume 119, 1992
688
Published November 1, 1992
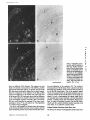
inin B2e and B2t mRNAs in
fetal lung. The B2e antisense
probe (A and B) shows strong
signals in endothelial cells of
arteries (a) and epithelial cells
of alveoli (al) but negligible
signalsin the bronchial (b) epithelium. The B2t probe (C and
D) shows strong signals both
in bronchial and alveolar epithelia, while the arteries are
negative. Bar, 110/zm.
tion and alkylation of the cysteines (35). The carboxylterminal portion of the Be chain which is absent in the short
B2t chain may, therefore, not be crucial for the formation of
an Ae-Ble-B2t heterotrimer. This is the sequence region
where an increased density of favorable interchain ionic interactions between the Ae and Ble chains can be found (not
shown). In such a conformation this region might form a
two-stranded rope structure similarly to several intracellular
proteins, including myosin and intermediate filament proteins. It cannot be excluded, however, that the B2t chain,
which lacks the carboxyl-terminal eysteine assembles with
some other, as yet unknown, laminin chains.
from two different cDNA libraries. This sequence is most
likely to be present in some portion of the mRNAs, possibly
generated by alternative splicing. At present, the part of the
B2t chain gene encoding this region has not been isolated.
However, comparison of the sequence of the B2t and B2e
chains and comparison to the known exon-intron structure
of the laminin B2e (38) chain gene shows that the difference
in the 3' end sequence is situated in the exon-intron junction
in the homologous sequence of the B2e chain. The shorter
B2t form could therefore be generated if the intron corresponding to intron 27 was not spliced out. This hypothesis
is currently being investigated.
For the in vitro assembly of laminin chains the presence
of the terminal disulfide bond may not be essential, since the
same reassembly products can be observed also after reduc-
The absence of domain VI distinguishes the B2t chain from
Kallunki et al. Novel Laminin B Chain
689
Unique Domain Structure of the Short Arm
Downloaded from on June 14, 2017
Figure 9. Expression of lam-
Published November 1, 1992
all other known laminin chains. This domain which forms
the terminal globules of the short arms has been shown to
participate in the association of laminin molecules in vitro
(8, 63) and recently it has been shown that laminin forms an
independent network in basement membranes in vivo (73).
Domain VI has also been reported to bind to type IV collagen (11, 42). The absence of domain VI from the B2t chain
indicates that it participates in another type of supramolecular structure than molecules containing the classical chains.
Another basement membrane protein, nidogen, has been
proposed to form a link between laminin and type IV collagen, binding to both proteins (23). The major binding site
for nidogen in laminin has been localized to EGF modules
in domain III of laminin B2e chain (28). In the laminin B2t
chain, most of the modules corresponding to the ones that
bind nidogen are missing from domain III. However, one of
the EGF modules in domain III which shows considerable
similarity to a module in domain III in the B2e chain is present. Therefore, it is possible that the B2t chain contains a
nidogen binding region. A synthetic decapeptide containing
a sequence from the carboxyl-terminal end of the mouse
laminin B2e chain has been reported to stimulate neurite outgrowth (43). The region containing this sequence is also
missing from the B2t chain. The smaller amount of potential
glycosylation sites in laminin B2t chain may also be of
significance as glycosylafion of laminin has been reported to
affect cell attachment, spreading, and neurite outgrowth (5,
16, 17). The binding sites for the integrin type cell surface
receptors have been localized to a region in the long ann
close to the terminal globule (27, 41, 67, 70a) and also to do-
The Journal of Cell Biology, Volume 119, 1992
690
Downloaded from on June 14, 2017
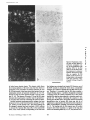
Figure 10. Expression of larninin B2e and B2t mRNAs in
embryonic metanephric kidney. In situ hybridization with
antisense RNA for the B2e
chain (A and B) shows abundant expression in embryonic
nephrons. The secretory nephrons are negative for B2t
mRNA (C and D) but the collecting tubules in the medulla
show clear expression. Bars:
(A and B) 110 txm; (C and D)
110/~m.
Published November 1, 1992
mains III (29). Recognition of different laminin isoforms by
different receptors could be of importance in a number of
biological processes. There are already data indicating differences in binding of cells to different laminin molecules
(7, 68).
Chromosomai Assignment of the B2t Chain Gene
This work was supported in part by grants from the Academy of Finland,
The Sigrid Jusetius Foundation and Finland's Cancer Institute (K. Tryggvason), The Finnish Cancer Foundation (H. Hirvonen), and by National Institutes of Health (NIH) grants HG00333 and HDO5196 (T. B. Shows).
Received for publication 11 June 1992 and in revised form 30 July 1992.
References
Previous studies have shown a general widespread distribution of the Ble and B2e chains, whereas the Bls chain is present primarily in synapses of motor neurons, renal glomeruli,
and arteries (58). The present study showed that the expression of the B2t chain gene differs from that of the Ble and
B2e genes regarding spatial expression in human fetal tissues. The Northern analyses confirmed previous observations (4, 40) showing slightly divergent expression of the Ble
and B2e genes in human tissues. However, the present results
suggest that although there are some differences in the levels
of Ble and B2e chain mRNAs, they appear to be expressed
in many tissues in a coordinated fashion.
The present in situ hybridization analyses with B2t and
B2e chain specific probes demonstrated that the corresponding genes are largely expressed in different cells in tissues
where both are expressed as shown by Northern analyses.
The major differences are that the B2e chain is expressed in
both epithelial cells and vascular endothelial, while expression of B2t is confined to epithelial cells. The particularly
strong expression of B2t in epithelial cells of adnexes in skin,
lung bronchi, as well as the collecting tubuli in kidney indicates that the B2t chain is associated with specialized basement membranes. The epithelial ceils in these regions are
characterized by their secretory function. Recently two proteins specific for epithelial basement membranes have been
isolated from keratinocyte cultures. Epiligrin has been localized by immunostaining of skin to the basement membranes
1. Barlow, D. P., N. M. Green, M. Kurkinen, and B. L M. Hogaru 1984.
Sequencing of laminin B chain cDNAa reveals C-terminal regions of
coiled-coil alpha-helix. EMBO (Fur. Mot. Biol. Organ.) J. 3:2355-2362.
2. Beck, K., I. Hunter, and J. Engel. 1990. Structure and function of lamiain:
anatomy of a multidomain glycoprotein. FASEB (Fed. Am. Soc. Erp.
Biol.) J. 4:148-160.
3. Beck, K., J. Spring, R. Chiquet-Ehrismann, J. Engel, and M. Chiquet.
1991. Structure of the basement membrane protein laminin: variations on
a theme. In Springer Series in Biophysics, Vol. 7. W. Taylor and P.
Argos, editors. Springer Verlag, Heidelberg/New York. 231-256.
4. Boot-Handford, R. P., M. Kurkinen, and D. J. Prockop. 1987. Steady-state
levels of mRNAs coding for the type IV collagen and laminin polypeptide
chains of basement membranes exhibit marked tissue-specific stoichiometric variations in the rat. J. Biol. Chem. 262:12475-12478.
5. Bouzon, M., C. Dussert, J. C. Lissitzky, and P. M. Martin. 1990. Spreading of B16F1 cells on laminin and its proteolytic fragments P1 and EB:
involvement of laminin carbohydrate chains. Exp. Cell Res. 190:47-56.
6. Boyd, C. D., K. Weliky, S. Toth-Fejel, S. B. Deak, A. M. Christiano,
J, W. MacKenzie, L. I. Sandell, K. Tryggvason, and E. Magenis. 1986.
The single copy gene coding for human ct I(IV) procollagen is located at
the terminal end of the long arm of chromosome 13. Hum. Genet. 74:
121-125.
7. Brown, J. C., and S. L. Goodman. 1991. Different cellular receptors for
human placental laminin and murine EHS laminin. FEBS (Fed. Eur. Biochem. Soc.) Lett. 282:5-8.
8. Bruch, M., R. Landwehr, and J. Engel. 1989. Dissection of laminin by
cathepsin G into its long-arm and short-arm structures and localization
of regions involved in calcium dependent stabilization and self-association. Fur. J. Biochem. 185:271-279.
9. Carter, W. G., M. C. Ryan, and P. J. Gahr. 1991. Epiligrin, a new cell
adhesion ligand for integrin a3/~l in epithelial basement membranes.
Cell. 65:599-610.
10. Cavener, D. R., and S. C. Ray. 1991. Eukaryotic start and stop translation
sites. Nucleic Acids Res. 19:3185-3192.
11. Charonis, A. S., E. C. Tsilibary, P. D. Yurchenco, and H. Furthmayr.
1985. Binding of laminin to type IV collagen: a morphological study. J.
Biol. Chem. 100:1848-1853.
12. Chi, H.-C., and C.-F. Hui. 1989. Primary structure of the Drosophila laminin 132 chain and comparison with human, mouse, and Drosophila laminin B1 and B2 chains. J. Biol. Chem. 264:1543-1550.
13. Cooper, A. R., and H. A. MacQueen. 1983. Subunits of laminin are
differentially synthesized in mouse eggs and early embryo. Dev. Biol. 96:
467-471.
14. Cooper, A. R., M. Kurkinen, A. Taylor, and B. L. M. Hogan. 1981.
Studies on the biosynthesis of laminin by murine parietal endoderm cells.
Eur. J. Biochem. 119:189-197.
Kallunki et al. Novel Laminin B Chain
691
Spatial Expression of B2t and B2e Chain mRNAs
Downloaded from on June 14, 2017
The high degree of sequence similarity between the B2t and
B2e chains suggests that the genes for these chains have
formed through duplication. This idea is supported by the
finding that the genes for these subunits are located close to
each other on chromosome 1. The gene for the laminin B2t
chain was localized in this study to 1q25~q31. Previously,
the gene for the laminin B2e chain was localized to the same
region (25). However, in the case of the laminin B2e chain
gene, the majority of signals localized over lq25, whereas for
the laminin B2t chain, the majority of signals localized over
lq31. The human genes for the basement membrane collagen
chains, od(IV) and c~2(IV) chains have been localized close
to each other on chromosome 13 (6, 30) and they have been
shown to have a common bidirectional promoter region and
transcription from opposite strands (55, 66). This is, however, not the case with the genes for the laminin B2e and B2t
chains, which have separate promoters and are probably
situated much further apart from each other (T. Kallunki,
unpublished data). At the present, the complete exon-intron
structure has been elucidated only for the human laminin Ble
(71) and B2e chains (38). Although these proteins have a
conserved domain structure, the structure of their genes
shows extensive divergence, with different exon-intron junctions that do not usually follow the boundaries of structural
domains or internal repeats.
of epidermis, sweat glands, and ducts and in lung to the basement membranes of ciliated epithelial cells and submucosal
glands in the bronchus (9). Epiligrin colocalizes with integrins c~3B1 and o~6B4 in focal adhesion contacts and stable adhesion contacts (hemidesmosomes). The molecular
masses of the subunits of epiligrin have been estimated to be
135, 145, and 170 kD. Kalinin is another protein isolated
from skin and keratinocyte cultures with almost same size
subunits and tissue localization in skin and lung in epithelial
basement membranes with hemidesmosomes (57). The exact relationship of these proteins has not been established.
It is possible that the B2t chain described in the present study
is related with or even a component of these proteins.
Together, the present and previous data emphasize the existence of a variety of laminin isoforms with distinct localization and possibly different biological functions. These data
also indicate that there are still unidentified laminin chains
and isoforms. Studies on the molecular assembly and biological functions of these molecules are a major challenge for
future studies.
Published November 1, 1992
bryonic mouse organs. Development (Camb.). 110:823-837.
40. Kleinman, H. K., I. Ebihara, P. D. Killen, M. Sasari, F. B. Cannon, Y.
Yamada, and G. R. Martin. 1987. Genes for basement membrane proteins are coordinately expressed in differentiating F9 cells but not in normal adult murine tissues. Dev. BioL 122:273-378.
41. Kramer, R. H., M. P. Vu, Y.-F. Cheng, D. M. Ramos, R. Timpl, and N.
Waleh. 1991. Laminin-binding integrin ce7Bl: functional characterization and expression in normal and malignant melanocytes. Cell Regul.
2:805-817.
42. Laurie, G. W., J. T. Bing, H. K. Kleinman, J. R. Hassell, M. AumaiUey,
G. R. Martin, and R. J. Feldmann. 1986. Localization of binding sites
for laminin, heparan sulfate proteoglyean and fibronectin on basement
membrane (type IV) collagen. J. Mol. Biol. 189:205-216.
43. Liesi, P., A. N~irv~inen,J. Soos, H. Sariola, and G. Snounou. 1989. Identification of a neurite outgrowth-promoting domain of laminin using synthetic peptides. FEBS (Fed. Eur. Biochem. SOc.) Lett. 244:141-148.
44. Maniatis, T., E. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A
Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
45. Deleted in proof.
46. McLachlan, A. D., and M. Steward. 1975. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. Biochem. J. 276:369379.
47. Mecham, R. P. 1991. Receptors for laminin on mammalian cells. FASEB
(Fed. Am. SOc. Exp. Biol.)J. 5:253g-2546.
48. Montell, D. J., and C. S. Goodman. 1988. Drosophila substrate adhesion
molecule: sequence of laminin B1 chain reveals donmins of homology
with mouse. Cell. 53:463-473.
49. Montell, D. J., and C. S. Goodman. 1989. Drosophila laminin: sequence
of B2 subunit and expression of all three subunits during embryogenesis.
J. Ceil BioL 109:2441-2453.
50. Nakai, H., M. G. Byers, T. B. Shows, and R. T. Taggart. 1986. Assignment of the pepsinogen gene complex (PGA) to human chromosome region 1lq13 by in situ hybridization. Cytogenet. Cell Genet. 43:215-217.
51. Nissinen, M., R. Vuolteenaho, R. Boot-Handford, P. Kallanki, and K.
Tryggvason. 1991. Primary structure of the human laminin A chain.
Limited expression in human tissues. Biochem. J. 276:369-379.
52. Paulsson, M., R. Deutzmann, R. Timpl, D. Dalzoppo, E. Odermatt, and
J. Engel. 1985. Evidence for coiled-coil a-helical regions in the long arm
of laminin. EMBO (Eur. Mol. Biol. Organ.) J. 4:309-316.
53. Pikkarainen, T., R. Eddy, Y. Fukushima, M. Byers, T. Shows, T. Pihlajaniemi, M. Saraste, and K. Tryggvason. 1987. Human luminia BI
chain, a multidomain protein with gene (iamBl) locus in the q22 region
of chromosome 7. J. Biol. Chem. 262:10454-10462.
54. Pikkarainen, T., T. Kallunki, and K. Tryggvason, 1988. Human laminin
B2 chain: comparison of the complete amino acid sequence with the B1
chain reveals variability in sequence homology between different structural domains. J. Biol. Chem. 263:6751-6758.
55. P6schl, E., R. Pollner, and K. Kfihn. 1988. The genes for the or(IV) and
et2(iv) chains of human basement membrane collagen type IV are arranged head-to-head and separated by a bidirectional promoter of unique
structure. EMBO (Eur. Mol. Biol. Organ.) J. 7:2687-2695.
56. Rentrop, M., B. Knapp, H. Winter, and J. Schweizer. 1986. Aminoalkylsilane treated glass slides as support for in situ hybridization of keratin
cDNAs to frozen tissue sections under varying fixation pretreatment conditions, Histochemistry. 18:271-276.
57. Rouselle, P., G. P. Lunstrum, D. R. Keene, and R. E. Burgeson. 1991.
Kaliniu: an epithelium-specific basement membrane adhesion molecule
that is a component of anchoring filaments. J. Cell Biol. t 14:565-576.
58. Sanes, J. R., E. EngvaU, R. Butkowski, and D. D. Hunter. 1990. Molecular heterogeneity of basal laminae: isoforms of lamiuin and collagen IV
at the neuromuscular junction and elsewhere. J. Cell Biol. 111:16851699.
59. Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with
chain-terminatinginhibitors. Proc. Natl. Acad. Sci. USA. 74:5463-5467.
60. Sasaki, M., and Y. Yamada. 1987. The laminin 132chain has a multidomain
structure homologous to the BI chain. J. Biol. Chem. 262:17111-17117.
61. Sasaki, M., S. Kato, K. Kohno, G. R. Martin, and Y. Yamada. 1987. Sequence of cDNA encoding the laminin BI chain reveals a multidomain
protein containing cysteine-rich repeats. Proc. Natl. Acad. Sci. USA. 84:
935-939.
62. Sasaki, M., H. K. Kleinman, H. Huber, R. Dentzmann, and Y. Yamada.
1988. Laminin, a multidomain protein. The A chain has a unique globular
domain and homology with the basement membrane proteoglycan and the
laminin B chains. J. Biol. Chem. 263:16536-16544.
63. Schittny, J. C., and P~ D. Yurchenco. 1990. Terminal short arm domains
of basement membrane laminin are critical for its self-assembly. J. Cell
Biol. 110:825-832.
64. Shows, T. B., A. Y. Sakaguchi, and S. L. Naylor. 1982. Mapping the human genome, cloned genes, DNA polymorphisms, and inherited disease.
In Advances in Human Genetics, Vol. 12. H. Harris and K. Hirschoru,
editors. Plenum Press, New York/London. 34t-452.
65. Shows, T., R. Eddy, L. Haley, M. Byers, M. Henry, T. Fujita, H. Matsui,
and T. Taniguchi. 1984. Interleukin 2 (IL-2) is assigned to human chromosome 4. Somatic Cell MoL Genet. 10:315-318.
The Journal of Cell Biology, Volume 119, 1992
692
Downloaded from on June 14, 2017
15. Cox, K. H., D. V. DeLeon, L. M. Angerer, and R. C. Angerer. 1984. Detection of mRNAs in sea urchin embryos by in situ hybridization using
asymmetric RNA probes. Dev. Biol. 101:485-502.
16. Dean, J. W., S. Chandrasekaren, and M. L. Tanzer. 1990. A biological
role of the carbohydrate moieties of laminin. J. Biol. Chem. 265:1255312556.
17. Dennis, J. W., C. A. Waller, and V. Schirrmacher. 1984. Identification
of asparagine-linked oligosaccharides involved in tumor cell adhesion to
laminin and type IV collagen. J. Cell Biol. 99:1416-1423.
18. Ehrig, K., I. Leivo, W. S. Argraves, E. Ruoslahti, and E. EngvalL 1990.
Merosin, a tissue-specific basement membrane protein, is a laminin-like
protein. Proc. Natl. Acad. ScL USA. 87:3264-3268.
19. Ekblom, M., G. Klein, G. Murgraner, L. Peeker, R. Deutzmann, R.
Timpl, and P. Ekblom. 1990. Transient and locally restricted expression
of laminin A chain mRNA by developing epithelial cells during kidney
organogenesis. Cell. 60:337-346.
20. Engel, J., I. Hunter, T. Sehnlthess, K. Beck, T. W. Dixon, and D. A. D.
Parry. 1991. Assembly of laminin isoforms by triple and double stranded
coiled-coil structures. Biochem. SOc. Trans. 19:839-844.
21. Engvall, E,. D. Earwicker, T. Haaparanta, E. Ruoslahti, andL R. Sanes.
1990. Distribution and isolation of four laminin variants tissue restricted
distribution of heterotrimers assembled from five different subunits. Cell
Regul. 1:731-740.
22. Fan, Y.-S., L. M. Davis, and T. B. Shows. 1990. Mapping small DNA
sequences by fluorescence in situ hybridization directly and banded
metaphase chromosomes. Proc. Natl. Acad. Sci. USA. 87:6223-6227.
23. Fox, J. W., U. Mayer, R. Nischt, M. Aumailley, D. Reinhardt, H. Wiedemann, K. Mann, R. Timpl, T. Krieg, J. Engel, and M.-L. Chu. 1991.
Recombinant nidogen consists of three globular domains and mediates
binding of iaminin to collagen type IV. EMBO (Eur. Mol. BioL Organ.)
J. 10:3137-3146.
24. Frenette, G. P., R. W. Ruddon, R. F. Krzesicki, J. A. Naser, and B. P.
Peters. 1989. Biosynthesis and deposition of a nonconvalent lamininheparan sulfate proteoglycan complex and other basal lamina components
by a human malignant cell line. J. Biol. Chem. 264:3078-3088.
25. Fukushima, Y., T. Pikkarainen, T. Kallunki, R. L. Eddy, M. G. Byers,
L. L. Hayley, W. M. Henry, K. Tryggvason, and T. B. Shows. 1988.
Isolation of a human laminin B2 (LAMB2) eDNA clone and assignment
of the gene to chromosome region lq25~q31. Cytogenetics Cell Genet.
48:137-141.
26. Garrison, K., A. J. MacKrell, and L H. Fessler. 1991. Drosophila laminin
A chain sequence, interspecies comparison, and domain structure of a
major carboxyl portion. J. Biol. Chem. 266:22899-22904.
27. Gehlsen, K. R., K. Dickerson, W. S. Argraves. E. Engvall, and E. Ruoslahti. 1989. Subunit structure of a laminin-binding integrin and localization of its binding site on lamiuin. J. Biol. Chem. 264:19034-19038.
28. Gerl, M., K. Mann, M. Aumailley, and R. Timpl. 1991. Localization of
a major nidogen-binding site to domain HI of laminin B2 chain. Eur. J.
Biochem, 202:167-174.
29. Goodman, S. L., M. Aumallley, and H. yon der Mark. 1991. Multiple cell
surface receptors for the short arms of laminin: cel#l integrin and RGDdependent proteins mediate cell attachment only to domains HI in murine
tumor laminin. J. Celt Biol. 113:931-941.
30. Griffin, C. A., B. S. Emmanuel, J. R. Hansen, W. K. Cavenee, and J. C.
Myers. 1987. Human collagen genes encoding basement membrane
, 1 (IV) and ~t2(IV) chains map to the distal long arm of chromosome 13~
Proc. Natl, Acad. Sci. USA. 84:512-516.
31. Haaparanta, T., L Uitto, E. Ruoslahti, and E. Engvall. 1991. Molecular
cloning of the eDNA encoding human laminin A chain. Matrix. I 1:
151-160.
32. Deleted in proof.
33. Heremans, A., B. van der Schueren, B. De Cock, M. Panlsson, J. Cassiman, H. G. van der Berghe, and G. David. 1989. Matrix-associated heparan sulfate proteoglycan: core protein-specific monoclonal antibodies
decorate the pericellular matrix of eormective tissue cells and the stromal
side of basement membranes. J. Cell Biol. 109:3199-3211.
34. Hunter, D. D., V. Shah, J. P. Merlie, and J. R. Sanes. 1989. A laminin-like
adhesive protein concentrated in the synaptic cleft of the neuromuscular
junction. Nature (Loud.). 338:229-234.
35. Hunter, I., T. Schulthess, M. Bruch, K. Beck, and J. Engel. 1990. Evidence for a specific mechanism of laminin assembly. Fur. J. Biochem.
188:205-211.
36. Hunter, I., T. Schulthess, and J, Engel. 1992. Laminin chain assembly by
triple and double stranded coiled-coil structures. J. Biol. Chem. 267:
6006-6011.
37. Kallunki, P., R. L. Eddy, M. G. Byers, M. Kestil~, T. B. Shows, and K.
Tryggvason. 1991. Cloning of human heparan sulfate proteoglycan core
protein assignment of the geue (HSPGI) to Ip36.1-'*p35 and identification of a BamHI restriction fragment length polymorphism. Genomics.
11:389-396.
38. Kallunki, T., J. Ikonen, L. Chow, P. Kallunki, and K. Tryggvason. 1991.
Structure of the human laminin B2 chain gene reveals extensive divergenee from the laminin B1 chain gene. J. BioL Chem. 266:221-228.
39. Klein, G., M. Ekblom, L. Fecker, R. Timpl, and P. Ekbiom. 1990. Differentiai expression of laminin A and B chains during development of era-
Published November 1, 1992
mann, and S. L. Goodman. 1991. Skeletal myoblasts utilize a novel
/31-series integrin and not o~6/31 for binding to the E8 and T8 fragments
of laminin. J. Biol. Chem. 266:23593-23601.
70b. yon Hejne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690.
71. Vuolteenaho, R., L. T. Chow, and K. Tryggvason. 1990. Structure of the
human laminin B1 chain gene. J. Biol. Chem. 265:15611-15616.
72. Wilkinson, D. G., and J. Green. 1990. Postimplantation Mammalian Embryos: A Practical Approach. A. J. Copp and D. L. Cockroft, editors.
IRL Press Oxford. 155-171.
73. Yurchenco, P. D., Y.-S. Cheng, and H. Colognato. 1992. Laminin forms
an independent network in basement membranes. J. Cell Biol. 117:
1119-1133.
74. Zabel, B. U., S. L. Naylor, A. Y. Sakaguchi, G. I. Bell, andT. B. Shows.
1983. High-resolution chromosomal localization of human genes for
amylase, proopiomelanocortin, somatostatin, and a DNA fragment
(D3S1) by in situ hybridization. Proc. Natl. Acad. Sci. USA. 80:69326936.
Kallunki et al. Novel Laminin B Chain
693
Downloaded from on June 14, 2017
66. Soininen, R., M. Huotari, S. L. Hostikka, D. J. Prockop, and K. Tryggvason. 1988. The structural genes for ctl and a2 chains of hunmn type IV
collagen are divergently encoded on opposite DNA strands and have an
overlapping promoter region. J. Biol. Chem. 263:17217-17220.
67. Sonnenberg, A., C. J. T. Lindres, P. W. Modderman, C. H. Damsky, M.
Aumailley, and R. Timpl. 1990. Integrin recognition of different cellbinding fragments of laminin (P1, E3, EB) and evidence that ct6/31 but
not a6134 functions as a major receptor for fragment E8. J. Cell Biol.
110:2145-2155.
68. Sonnedherg, A., K. R. Gehlsen, M. Aumailley, and R. Timpl. 1991. Isolation of a6131 integrins from platelets and adherent cells by affinity chromatography on mouse laminin fragment E8 and human laminin pepsin
fragment. Exp. Cell Res. 197:234-244.
69. Timpl, R. 1989. Structure and biological activity of basement membrane
proteins. Fur. J. Biochem. 180:487-502.
70. Timpl, R., H. Rohde, P. Gehron Robey, S. I. Rennard, J. M. Foidart, and
R. R. Martin. 1979. Laminin-a glycoprotein from basement memhranes. J. Biol. Chem. 254:9933-9937.
70a. vonder Mark, H., J. Diirr, A. Sonnenberg, K. yon der Mark, R. Deutz-