* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Genetic Recombination in Eukaryotes
Human genome wikipedia , lookup
Epigenomics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
DNA vaccination wikipedia , lookup
Minimal genome wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Ridge (biology) wikipedia , lookup
Point mutation wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA supercoil wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Population genetics wikipedia , lookup
Neocentromere wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Microsatellite wikipedia , lookup
Genomic library wikipedia , lookup
Gene expression programming wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Holliday junction wikipedia , lookup
Genome editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
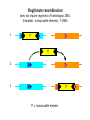
Genetic Recombination in Eukaryotes Message Recombinants are those products of meiosis with allelic combinations different from those of the haploid cells that formed the meiotic diploid. In meiosis, recombinant products with new combinations of parental alleles are generated by: 1. independent assortment (segregation) of alleles on nonhomologous chromosomes. 2. crossing-over in meiotic synaptonemal complexes between nonsister homologs. Meiosis in a diploid dihybrid cell. Genotype A/a; B/b The detection of recombination in diploid organisms. The advantage of a testcross (homozygote recessive tester) Independent assortment Two unlinked genes produce always a recombinant frequency of 50%. (Testcross of a dihybrid) Self of a dihybrid Prunett square showing the genotypic and phenotypic ratios. Genes Phenotypes Genotypes 1 2 3 2 4 9 3 8 27 . . . n 2n 3n Dihybrid selfing Cross between two A/a ; B/b dihybrids – recombination occurs in both members of cross – recombination frequency is 50% A;B A;b a;B a;b A;B A/A ; B/B A/A ; B/b A/a ; B/B A/a ; B/b A;b A/A ; B/b A/A ; b/b A/a ; B/b A/a ; b/b a;B A/a ; B/B A/a ; B/b a/a ; B/B a/a ; B/b a;b A/a ; B/b A/a ; b/b a/a ; B/b a/a ; b/b 9 A/– ; B/– 3 A/– ; b/b Ratio: 3 a/a ; B/– 1 a/a ; b/b 9:3:3:1 segregation in maize Crossing-over Chiasmata at meiosis. Each line represents a chromatid of a pair of synapsed chromosomes In dihybrids for linked genes, recombinants arise from meioses in which nonsister chromatids cross over between the genes under study. Recombinants produced by crossing-over Linkage Symbolism: A A B B a A b B Genetic Maps (linkage maps) Message Recombination between linked genes can be used to map their relative distance on the chromosome. The map unit (1m.u. or 1cM) is defined as a recombinant frequency of 1%. In a dihybrid of linked genes the RF will be between 0% and 50%. Recombination frequency (RF) • Experimentally determined from frequency of recombinant phenotypes in testcrosses • Roughly proportional to physical length of DNA between loci • Greater physical distance between two loci, greater chance of recombination by crossingover • 1% recombinants = 1 map unit (m.u.) • 1 m.u. = 1 centiMorgan (cM) Linkage maps # observed 140 50 60 150 • RF is (60+50)/400=27.5%, clearly less than 50% • Map is given by: A 27.5 m.u. B Mapping • RF analysis determines relative gene order • RF between same two loci may be different in different strains or sexes • RF values are roughly additive up to 50% – multiple crossovers essentially uncouple loci, mimicking independent assortment • Maps based on RF can be combined with molecular and cytological analyses to provide more precise locations of genes Genetic maps • Useful in understanding and experimenting with the genome of organisms • Available for many organisms in the literature and at Web sites • Maps based on RF are supplemented with maps based on molecular markers, segments of chromosomes with different nucleotide sequences Comparison of physical and genetic maps The yeast chromosome 1 is shown. A) indicates a region where the genetic map is contracted owing to decreased frequency of crossingover. B) indicates a region where the genetic map is expanded owing to increased frequency of crossingover. The mechanism of crossing-over Three types of DNA recombination: 1. Homologous recombination 2. Site-specific recombination 3. Illegitimate recombination The mechanism of crossing-over Two types of homologous recombination. Crossover between two dsDNA molecules results in the reciprocal exchange of DNA. Gene conversion involves a nonreciprocal transfer. The donor sequence remains unchanged, while the recipient sequence is changed. The Meselson-Radding heteroduplex model. a) b) c) d) single stranded nick DNA polymerase ssDNA displaces its counterpart in the homologue displaced ssDNA is digested The Meselson-Radding heteroduplex model. e) f) ligation completes the formation of a Holliday junction resolution according to Holliday model in two alternative ways creats either a crossover chromatid (V) or a non-crossover chromatid (H). Repair of mismatched nucleotides in heteroduplex DNA fungal tetrads for segregation analysis An A/a meiocyte undergoes meiosis, resulting in an equal number of A and a products. The abberant 5:3 octad is explained by a heteroduplex formed during meiosis. In this case the nucleotide differences in the heteroduplex are not repaired. The role of RecA in strand transfer. The E. coli RecA protein binds to ssDNA. The resulting nucleoprotein complex aggregates with dsDNA in a triplestranded DNA complex in which the bases do not pair. This complex facilitates invasion of the ssDNA. Strands are subsequently exchanged and a heteroduplex can be formed. Site-specific recombination involves defined DNA sites, is independent of RecA, and requires specific enzymes. (Examples: bacteriphage λ) Integration of λ DNA into the E. coli chromosome involves site-specific recombination between the attP sequence of the phage and the bacterial attB sequence. The recombination is catalysed by an integrase. Illegitimate recombination does not require segments of homologous DNA. (Examples: transposable elements, T-DNA) 1 T T 2 3 T T = transposable element Mitotic Crossing-over Message A mitotic crossover generates homozygosity of alleles of heterozygous loci distal to the crossover. Mitotic Recombination in Drosophila Cross: y+ sn / y+ sn X y sn+ / y sn+ Mitotic Recombination in Drosophila y sn+ / y sn y sn+ / y sn+ y+ sn / y+ sn