* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gill: Human Disease Genomics
Gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Transposable element wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Metagenomics wikipedia , lookup
Frameshift mutation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Point mutation wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Pathogenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genomic library wikipedia , lookup
Human genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Genome (book) wikipedia , lookup
Genome editing wikipedia , lookup
Human Genome Project wikipedia , lookup
Oncogenomics wikipedia , lookup
CS273A
Lecture 18: Human Disease Genomics
http://cs273a.stanford.edu [Bejerano Fall16/17]
1
Announcements
• Hope my vocal chords last till 2:50 :)
http://cs273a.stanford.edu [Bejerano Fall16/17]
2
START WITH HYPERBOLE
http://cs273a.stanford.edu [Bejerano Fall16/17]
3
The Human Genome
Genome
1013 cells
≈
Human
Genome = The Operating System that runs every cell in our body
3*109 letters long, over the DNA alphabet = {A,C,G,T}
http://cs273a.stanford.edu [Bejerano Fall16/17]
4
The Biggest Challenge in Genomics…
… is computational:
How does this
Program
encode this
Output
This “coding” question has profound implications for our lives
http://cs273a.stanford.edu [Bejerano Fall16/17]
5
Match to the Next Three Slides…
Time
Negative Selection
Neutral Drift
http://cs273a.stanford.edu [Bejerano Fall16/17]
Positive Selection
6
The Biggest Challenge in Genomics…
… is computational:
How does this
encode this
Program
Forks & re-merges
Where did we come from? How are we different from each other?
http://cs273a.stanford.edu [Bejerano Fall16/17]
7
The Biggest Challenge in Genomics…
… is computational:
How does this
encode this
Program
Suite of related products
What in our genomes make us different from other species?
http://cs273a.stanford.edu [Bejerano Fall16/17]
8
The Biggest Challenge in Genomics…
… is computational:
How does this
Program
encode this
Bugs
Output
What genomic mutations predispose us to disease?
http://cs273a.stanford.edu [Bejerano Fall16/17]
9
The Biggest Challenge in Genomics…
… is computational:
How does this
Program
encode this
Bugs
Patching
What genomic mutations determine our drug response?
http://cs273a.stanford.edu [Bejerano Fall16/17]
10
The Biggest Challenge in Genomics…
… is computational:
How does this
Program
encode this
Bugs
Verification
We can eliminate suffering by not “booting” “buggy” embryos
http://cs273a.stanford.edu [Bejerano Fall16/17]
11
The Biggest Challenge in Genomics…
… is computational:
How does this
Program
encode this
Bugs
Debugging
We can eliminate suffering by fixing people’s “buggy” genomes
http://cs273a.stanford.edu [Bejerano Fall16/17]
12
Literally Save Lives From Your Keyboard!
Read
Understand
Fix
http://cs273a.stanford.edu [Bejerano Fall16/17]
13
Gene Therapy: We Design the Cure
Life saving
“code injection”
http://cs273a.stanford.edu [Bejerano Fall16/17]
14
Biomedicine is facing a phase transition
From an obsession with the interpreter,
Code
Interpreter
Output
To an obsession with the code.
(When your code has a bug what do you fix?..)
http://cs273a.stanford.edu [Bejerano Fall16/17]
15
Gene Therapy 3.0: Precise, Hereditary
http://cs273a.stanford.edu [Bejerano Fall16/17]
16
Every New Technology Raises Ethical Issues
http://cs273a.stanford.edu [Bejerano Fall16/17]
17
Human disease
http://cs273a.stanford.edu [Bejerano Fall16/17]
18
Even single basepair mutations can be
devastating (to the individual)
The Species Tree
S
S
Sampled Genomes
S
Speciation
20
Time
Fixation, Positive & Negative Selection
Time
Negative Selection
Neutral Drift
http://cs273a.stanford.edu [Bejerano Fall16/17]
Positive Selection
21
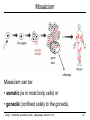
Mosaicism
germ
cells
offsprings
Mosaicism can be:
• somatic (ie in most body cells) or
• gonadal (confined solely to the gonads).
http://cs273a.stanford.edu [Bejerano Fall16/17]
22
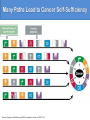
Fundamental Changes in Cancer Cell Physiology
Exploitation of natural pathways
for cellular growth
• Growth Signals (e.g. TGF family)
• Angiogenesis
• Tissue Invasion & Metastasis
Evasion of anti-cancer control
mechanisms
• Apoptosis (e.g. p53)
• Antigrowth signals (e.g. pRb)
• Cell Senescence
Acceleration of Cellular Evolution
Via Genome Instability
• DNA Repair
• DNA Polymerase
Hanahan and Weinberg. 2000. The hallmarks of cancer. Cell 100: 57-70.
Many Paths Lead to Cancer Self-Sufficiency
Hanahan, Douglas, and Ra Weinberg. 2000. The hallmarks of cancer. Cell 100: 57-70.
Cancer Mutations
The relation between sporadic and inherited forms of the same tumour.
The target tissue contains n cells and the chance of one cell suffering a loss of
function mutation in the tumour suppressor gene is µ.
http://cs273a.stanford.edu [Bejerano Fall16/17]
25
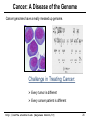
Cancer: A Disease of the Genome
Cancer genomes have a really messed up genome.
Challenge in Treating Cancer:
Every tumor is different
Every cancer patient is different
http://cs273a.stanford.edu [Bejerano Fall16/17]
26
Cancer Genomics Goal
http://cs273a.stanford.edu [Bejerano Fall16/17]
27
Statistical Genetics
http://cs273a.stanford.edu [Bejerano Fall16/17]
28
Genome Wide Association Studies (GWAS)
Control
A/G
A/G
G/G
G/G
A/G
G/G
G/G
Disease
A/A
A/G
A/A
A/G
A/G
A/A
A/A
AA
0
4
AG
3
3
GG
4
0
p-value
Genome Semantics
• The Genome is ultimately a Programming Language
• Statistical approaches ignore genome semantics
• Population Genetics (neutral drift)
• Selective sweeps (positive selection)
• GWAS (negative selection)
• All ask questions about allele transmission
• Not allele meaning
http://cs273a.stanford.edu [Bejerano Fall16/17]
30
Consumer Genomics
1 Collect scientific literature
about all structural variant
correlations with human
disease & traits.
2 Genotype customers for as
many informative loci as is
commercially viable.
3 Offer counseling for your
findings, and their meaning.
4 Ask customers to
phenotype themselves.
5 Discover new associations!
http://cs273a.stanford.edu [Bejerano Fall16/17]
Knowledge = technology + pre-conceptions
Our knowledge is strongly influenced by what our
technology allows us to see. What it does not, we will with
pre-conceptions. And those may often be wrong.
Even after half the human genome was sequenced, people
thought there will be over 100,000 coding genes in it.
There are in fact ~20,000 (in par with the gene repertoire of
much simpler animals).
Before the next generation sequencing revolution people
thought the majority (99%) of human disease mutations
will be in the coding regions.
80-90% of human disease GWAS associated SNPs are in
fact non-coding. The majority is likely gene regulatory.
http://cs273a.stanford.edu [Bejerano Fall16/17]
32
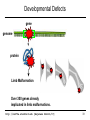
Developmental Defects
gene
genome
protein
Limb Malformation
Over 300 genes already
implicated in limb malformations.
http://cs273a.stanford.edu [Bejerano Fall16/17]
33
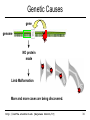
Genetic Causes
gene
genome
NO protein
made
Limb Malformation
More and more cases are being discovered.
http://cs273a.stanford.edu [Bejerano Fall16/17]
34
Modularity: Disease Implications
http://cs273a.stanford.edu [Bejerano Fall16/17]
35
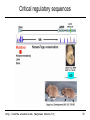
Critical regulatory sequences
http://cs273a.stanford.edu [Bejerano Fall16/17]
36
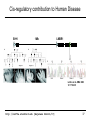
Cis-regulatory contribution to Human Disease
SHH
1Mb
LMBR1
Limb
Lettice et al. HMG 2003
12: 1725-35
http://cs273a.stanford.edu [Bejerano Fall16/17]
37
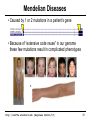
Mendelian Diseases
• Caused by 1 or 2 mutations in a patient’s gene
• Because of “extensive code reuse” in our genome
these few mutations result in complicated phenotypes
http://cs273a.stanford.edu [Bejerano Fall16/17]
38
Mendelian Diseases
• There are 8,000 known rare Mendelian diseases
• Each can cause over a dozen different phenotypes
of 10,000 known disease phenotypes
• Together rare Mendelian diseases affect 1 in 33 babies
• There are over 20,000 genes in the human genome
• Sequencing all genes is cheap, and getting cheaper
• We now know of thousands of genes that when mutated
cause thousands of different Mendelian diseases.
• Diagnosing a case is vital for patient & their family
• Because doctors rarely see each rare disease twice,
and because there are so many genes and so many
rare diseases, MANUAL DIAGNOSIS IS UNSUSTAINABLY
SLOW, INACCURATE AND EXPENSIVE.
http://cs273a.stanford.edu [Bejerano Fall16/17]
39
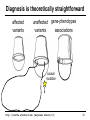
Diagnosis is theoretically straightforward
affected
variants
unaffected gene-phenotypes
associations
variants
causal
mutation
http://cs273a.stanford.edu [Bejerano Fall16/17]
40
WHAT NEXT?
http://cs273a.stanford.edu [Bejerano Fall16/17]
41
Beyond Coding
CODING
SNV
1-50bp
mutations
Exome
http://cs273a.stanford.edu [Bejerano Fall16/17]
42
Targeted Sequencing, or
looking under the lamp
Exome
Library
Shotgun
Library
Exon 1
Exon 2
Genomic DNA
Capture Methods vs. Shotgun
•
Targeted sequencing allows for
much higher coverage at less cost
• Will only capture known sites
• These methods also introduce
significant captures bias, including
0.1% SNP array $100 failure to capture sites that differ
significantly from the reference
2% Exome
$1000 genome. (analogous to microarrays)
100%* Genome $2000
http://cs273a.stanford.edu [Bejerano
Modified from Meyerson et al. . 2010. Advances in understanding cancer genomes through
Fall16/17]
second-generation sequencing. Nature Reviews Genetics 11, no. 10 (October): 685-696
Beyond Coding
CODING
SNV
1-50bp
mutations
SV
rest of
spectrum
NONCODING
Exome
http://cs273a.stanford.edu [Bejerano Fall16/17]
44
Beyond Coding
SNV
1-50bp
mutations
SV
rest of
spectrum
CODING
NONCODING
Exome
Genome
Genome
Genome
http://cs273a.stanford.edu [Bejerano Fall16/17]
45
http://cs273a.stanford.edu [Bejerano Fall16/17]
46