* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Presentasi 1
Survey
Document related concepts
Transcript
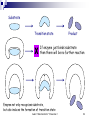
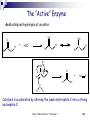
Basic enzyme Aulanni’am Biochemistry Laboratory Brawijaya University Aulani " Biokimia Enzim " Presentasi 1 1 What are enzymes ? Enzymes are proteins They have at least one active site Active site is lined with residues and sometimes contains a co-factor Active site residues have several important properties: Charge [partial, dipoles, helix dipole] pKa Hydrophobicity Flexibility Reactivity (Cysteines) Aulani " Biokimia Enzim " Presentasi 1 2 What are chemical reactions? In a chemical reactions a compound “A” is changed into a compound “B”. In context of biochemistry, chemical reactions are “organic chemistry reactions”. In organic chemistry reactions bonds are broken and/or formed (generalization) Bonds are “paired electrons” between two nuclei (C-C, C=C, C-O,C=O, C-H, O-H, N-H etc.) Thus reactions involve “rearranging” electrons In context of biochemistry, a frequent player in chemical reactions is H2O (hydronium H3O+ and hydroxide OH-) Aulani " Biokimia Enzim " Presentasi 1 3 Enzyme catalysis Enzyme catalysis is characterized by two features Substrate specificity Rate acceleration Aulani " Biokimia Enzim " Presentasi 1 4 Enzyme substrate specificity Unlike “chemical catalysts” enzyme only catalyze reactions for a “relatively” narrow substrate spectrum. For example: substrate spectrum of restriction enzymes, and protein kinases. Two main theories for substrate specificity Lock-and-Key hypothesis (Fisher, 1894) Induced-fit hypothesis (Koshland, 1958) Aulani " Biokimia Enzim " Presentasi 1 5 Substrate Transition state X Product If enzyme just binds substrate then there will be no further reaction Enzyme not only recognizes substrate, but also induces the formation of transition state Aulani " Biokimia Enzim " Presentasi 1 6 The Nature of Enzyme Catalysis ● Enzyme provides a catalytic surface ● This surface stabilizes transition state ● Transformed transition state to product B A A B Aulani " Biokimia Enzim " Presentasi 1 Catalytic surface 7 Lock-and-Key vs. Induced-Fit Lock-and-Key does not always explain substrate spectrum (e.g. analogs smaller than substrate don’t bind while analogs larger than substrate do bind) Induced-fit implies the concepts: conformational change catalytically competent conformation (low catalytic form and high catalytic form) Aulani " Biokimia Enzim " Presentasi 1 8 Catalyzed vs. un-catalyzed reactions Free Energy (delta G) S‡ S‡c ES‡ S ES EP P Reaction Coordinate Aulani " Biokimia Enzim " Presentasi 1 9 Specific Induced to transition state + N H N H = O C H +d + Acid catalysis C H N H = N H C O H H H -d O H O C H = N H Base catalysis Fast C H O H H Slow O C H O H - O O C H = Acid-base Catalysis Both N C H O H H H O - O C C Fast Very Fast Aulani " Biokimia Enzim " Presentasi 1 10 Rate Acceleration Catalyzes of a reaction results in rate enhancement not alteration of the equilibrium Catalysis involves reduction of activation energy This can be most readily done by lowering the Free Energy of the transition state Additionally the Free Energy of the ground state can be raised (not a general strategy) Free Energy (delta G) S‡ ES‡ S ES EP P Reaction Coordinate Aulani " Biokimia Enzim " Presentasi 1 11 Transition state Stabilization by Enzyme S‡ Free Energy (delta G) How does an Enzyme reduce the Activation Energy ?? Enzyme stabilizes the transition state, i.e. makes the “strained” conformation more bearable. Note: An enzyme can only reduce the activation energy if it binds better to the transition state than to the substrate [otherwise, the DDG between ES and ES‡ is the same as between S and S‡] Aulani " Biokimia Enzim " Presentasi 1 ES‡ S ES EP P Reaction Coordinate 12 Transition state Stabilization by Enzyme Compounds that closely mimic the transition state bind much better to an enzyme than the original substrate. Transition state analogs are potent inhibitors (pico molar affinities) Free Energy (delta G) Implications of preferential stabilization of the transition state. S‡ ES‡ S ES EP P Reaction Coordinate Applications: • Inhibitor/drug development based on transition state model • Development of catalytic antibodies [rate acceleration up to 105] Aulani " Biokimia Enzim " Presentasi 1 13 Enzyme Stabilizes Transition State Energy change EST S ES EP P Energy decreases (under catalysis) Energy required (no catalysis) T = Transition state ST Reaction direction What’s the difference? Aulani " Biokimia Enzim " Presentasi 1 14 Active Site Is a Deep Buried Pocket Why energy required to reach transition state is lower in the active site? It is a magic pocket (1) Stabilizes transition + CoE (1) (4) - (2) Expels water (2) (3) Reactive groups (4) Coenzyme helps (3) Aulani " Biokimia Enzim " Presentasi 1 15 Enzyme Active Site Is Deeper than Ab Binding Ag binding site on Ab binds to Ag complementally, no further reaction occurs. Instead, active site on enzyme also recognizes substrate, but actually complementally fits the transition state and stabilized it. X Aulani " Biokimia Enzim " Presentasi 1 16 Enzyme mediated catalysis Strategies for transition state stabilization and/or ground state destabilization: Proximity Strain or distortion Orbital steering However, additionally the enzyme can be an “active” participant in reaction Acid/base catalysis Nucleophilic/electrophilic catalysis Covalent catalysis Aulani " Biokimia Enzim " Presentasi 1 17 Rate Acceleration: Proximity For un-catalyzed reactions involving two substrates the rate can be increased by increasing the number of collisions (higher temperature) Enzymes capture each substrate (sometimes in a ordered manner) and appropriately orient them with respect to each other, thus obviating the need for higher temperature The capture of substrates by the enzyme has an Entropic cost; this cost must be compensated by favourable interactions between enzyme and substrates The effect of confining the substrates in the active site of the enzyme is similar to raising the concentration of the substrates. Hence, the proximity effect is also referred to as increasing the effective concentration Aulani " Biokimia Enzim " Presentasi 1 18 Active Site Avoids the Influence of Water + Preventing the influence of water sustains the formation of stable ionic bonds Aulani " Biokimia Enzim " Presentasi 1 19 Essential of Enzyme Kinetics Steady State Theory E + S E S E +P In steady state, the production and consumption of the transition state proceed at the same rate. So the concentration of transition state keeps a constant. Aulani " Biokimia Enzim " Presentasi 1 20 Constant ES Concentration at Steady State S P Concentration E ES Reaction Time Aulani " Biokimia Enzim " Presentasi 1 21 The “Active” Enzyme Examine the hydrolysis of an ester: O R' R O very slow + + H2O O Weak electrophile R R' OH Poor nucleophile O O Expected transition state HO R' R O H R' R O O H O d d H H Aulani " Biokimia Enzim " Presentasi 1 22 The “Active” Enzyme Base catalyzed hydrolysis of an ester: O O O R' R O + R - O + R' OH R HO O- OH Catalysis is accelerated by altering the poor nucleophile H2O into a strong nucleophile OH- Aulani " Biokimia Enzim " Presentasi 1 23 R' The “Active” Enzyme Acid catalyzed hydrolysis of an ester: + OH O R' R + H3O+ OH R' R O C+ O R R' O d OH C R d O R' + O R HO R' OH O H H Catalysis is accelerated by altering the weak electrophile C into a strong nucleophile C+ Aulani " Biokimia Enzim " Presentasi 1 24 The “Active” Enzyme In standard organic chemistry for ester hydrolysis one has to choose between base or acid catalysis In enzyme catalysis the reaction is “carried out on a solid support” As a consequence one can incorporate both acid and base catalysis: Aulani " Biokimia Enzim " Presentasi 1 25 The “Active” Enzyme Enzyme catalyzed hydrolysis of an ester: B+ H Active site incorporates both: O R' R • a base [-B:] O O H H B: • an acid [-B+-H] B: B+ B: H d OH O C R' R R O H B: H B+ H d R' C O - O O HO R O O H Aulani " Biokimia Enzim " Presentasi 1 B+ H H 26 R' Catalysis of Phosphorylation Phosphorylation a very frequent reaction (e.g. signal transduction) Phosphoryl donating group is generally a nucleotide, e.g. ATP, GTP Transfer of phosphoryl group to: Water : hydrolysis [ATPase, GTPase] Anything else: phosphorylation [Kinase] Aulani " Biokimia Enzim " Presentasi 1 27 Mechanisms of Enzyme Catalyzed Phosphorylation Several mechanism are observed in Nature Reactions with covalent enzyme intermediates Direct inline transfer Perhaps metal assisted mechanisms Present two examples: Aminoglycoside kinases (Cousin of Protein kinases) G-proteins Aulani " Biokimia Enzim " Presentasi 1 28 An Example for Enzyme Kinetics (Invertase) 1) Use predefined amount of Enzyme →E → S (x 2) Add substrate in various concentrations 3) Measure Product in fixed Time (P/t) → vo (y 4) (x, y) plot get hyperbolic curve, estimate → Vmax 5) When y = 1/2 Vmax calculate x ([S]) → Km Vmax 1 vo -1 Km vo 1/2 1 Vmax Double reciprocal 1/S Aulani " Biokimia Enzim " Presentasi 1 Km Direct plot S 29 A Real Example for Enzyme Kinetics Data Substrate Product Velocity Double reciprocal [S] Absorbance v (mmole/min) 0.25 0.21 → 0.42 0.50 0.36 → 0.72 1.0 0.40 → 0.80 2.0 0.46 → 0.92 no 1 2 3 4 1/S 4 2 1 0.5 1/v 2.08 1.56 1.35 1.16 Double reciprocal Direct plot (1) The product was measured by spectroscopy at 600 nm for 0.05 per mmole (2) Reaction time was 10 min 1.0 v 0.5 0 0 1 2 [S] 2.0 1.0 1/v 1.0 -3.8 0 -4 Aulani " Biokimia Enzim " Presentasi 1 -2 0 2 1/[S] 4 30 Enzyme Inhibition (Mechanism) Equation and Description Cartoon Guide I Competitive I Non-competitive Substrate E S S E I Compete for Inhibitor active site S I I Uncompetitive S E I I Different site E + S← → ES → E + P + I ↓↑ EI E + S← → ES → E + P + + I I ↓↑ ↓↑ EI + S →EIS [I] binds to free [E] only, and competes with [S]; increasing [S] overcomes Inhibition by [I]. [I] binds to free [E] or [ES] complex; Increasing [S] can not overcome [I] inhibition. Aulani " Biokimia Enzim " Presentasi 1 S I E + S← → ES → E + P + I ↓↑ EIS [I] binds to [ES] complex only, increasing [S] favors the inhibition by [I]. 31 Enzyme Inhibition (Plots) I Competitive I Non-competitive Direct Plots Vmax vo vo I Double Reciprocal Km Km’ I [S], mM Km = Km’ I Uncompetitive Vmax Vmax Vmax’ Vmax’ [S], mM I Km’ Km [S], mM Vmax unchanged Km increased Vmax decreased Km unchanged Both Vmax & Km decreased 1/vo 1/vo 1/vo Intersect at Y axis 1/Km I I I Two parallel lines 1/ Vmax 1/[S] Intersect at X axis 1/Km 1/ Vmax 1/[S] Aulani " Biokimia Enzim " Presentasi 1 1/ Vmax 1/Km 1/[S] 32 H = O C–O - H–N C N H–O–CH2 C C H CH2 Asp 102 Ser 195 His 57 Active Ser H = O C–O–H Asp 102 N C N–H - O–CH C C H CH2 2 Ser 195 His 57 Aulani " Biokimia Enzim " Presentasi 1 33 pH Influences Chymotrypsin Activity Relative Activity 5 6 7 8 9 10 11 pH Aulani " Biokimia Enzim " Presentasi 1 34 Buffer pH pH Influences Net Charge of Protein 10 9 8 7 Isoelectric point, pI + + 6 5 4 3 0 Net Charge of a Protein Aulani " Biokimia Enzim " Presentasi 1 35 Imidazole on Histidine Is Affected by pH pH 6 H H–N C C + N–H H+ H–N C C H = Asp 102 H–N C C N C H H O C–O - pH 7 H Inactive + N–H H–O–CH2 C C-H CH2 Ser 195 His 57 Aulani " Biokimia Enzim " Presentasi 1 36 Chymotrypsin Produces New Ile16 N-Terminal New NH2-terminus Relative activity L13 pH 5 6 7 8 I16 Y146 9 10 11 NH2– Ile 16 Asp 194 pH 9 –CH2COO+ NH 3– pKa pH 10 Ile 16 Aulani " Biokimia Enzim " Presentasi 1 37 New Ile16 N-Terminal Stabilizes Asp194 Catalytic Triad His 57 Asp 102 Ser 195 Gly 193 Asp 194 +NH 3 Ile 16 Aulani " Biokimia Enzim " Presentasi 1 38 Chymotrypsin Ser195 Inhibited by DIFP Diisopropyl-fluorophosphate (DIFP) X = O (CH3)2CH–O– P –O–CH(CH3)2 F = O (CH3)2CH–O– P –O–CH(CH3)2 O-…H O CH2 CH2 Ser 195 Ser 195 Aulani " Biokimia Enzim " Presentasi 1 39 Addition of Substrate Blocks DIFP Inhibition 100 No substrate Percent Inhibition of activity (%) + DIFP X 50 + DIFP & substrate Add substrate S 0 Reaction time Aulani " Biokimia Enzim " Presentasi 1 40 Chymotrypsin Also Catalyzes Acetate Hartley & Kilby O -C NH Nitrophenol acetate O CH3–C–O– + H 2O –NO2 Chymotrypsin Peptide bond O -C OEster bond HO– –NO2 Acetate O CH3–C–OH Nitrophenol No acetate was detected at early stage Aulani " Biokimia Enzim " Presentasi 1 41 Two-Stage Catalysis of Chymotrypsin O Nitrophenol acetate CH3–C–O– –NO2 Acylation O CH3–C –NO2 O C O-H C + H2O Aulani " Biokimia Enzim " Presentasi 1 Nitrophenol CH3COOH Deacylation (slow step) Kinetics of reaction OC HO– Two-phase reaction Time (sec) 42 Extra Negative Charge Was Neutralized -C-C-N-C-C-N-C-C-NH H O-C NHO H O-C NHO H O -C NH E+S O -C-OH NH2- Aulani " Biokimia Enzim " Presentasi 1 43 Active Site Stabilizes Transition State Gly 193 Catalytic Triad Ser 195 Asp 194 Met 192 His 57 Active Site Cys 191 Asp 102 Thr 219 Cys 220 Specificity Site Ser 214 Trp 215 Gly 216 Ser 218 Ser 217 Aulani " Biokimia Enzim " Presentasi 1 44 Regulation of Enzyme Activity Inhibitor Proteolysis or o I x I I S proteolysis x I o S inhibitor Feedback regulation R o Phosophorylation P x x o S R S regulator effector (+) P phosphorylation Signal transduction x (-) A or Regulatory subunit o S + Aulani " Biokimia Enzim " Presentasi 1 A cAMP or calmodulin 45 Classification of Proteases Family Metal Protease Serine Protease Cysteine Protease Aspartyl Protease Example Carboxypeptidase A Chymotrypsin Trypsin Mechanism Zn Specificity Inhibitor 2+ E72 H69 H196 S195-OH57 D102 C25-S- Nonpolar EDTA EGTA Aromatic Basic DFP TLCK TPCK PCMB Leupeptin Pepstatin Papain H195 Nonspecific Pepsin Renin D215 H2O D32 Nonspecific Aulani " Biokimia Enzim " Presentasi 1 46 Ser 195 Chymotrypsin Trypsin Elastase Thrombin Plasmin Acetylcholinesterase – Gly – Asp – Ser – Gly – Gly – Pro – Leu – – Gly – Asp – Ser – Gly – Gly – Pro – Val – – Gly – Asp – Ser – Gly – Gly – Pro – Leu – – Gly – Asp – Ser – Gly – Gly – Pro – Phe – – Gly – Asp – Ser – Gly – Gly – Pro – Leu – – Gly – Glu – Ser – Ala – Gly – Gly – Ala – His 57 Chymotrypsin Trypsin Elastase Thrombin Plasmin Acetylcholinesterase – Val – Thr – Ala – Ala – His – Cys – Gly – – Val – Ser – Ala – Gly – His – Cys – Tyr – – Leu – Thr – Ala – Ala – His – Cys – Ile – – Leu – Thr – Ala – Ala – His – Cys – Leu – – Leu – Thr – Ala – Ala – His – Cys – Leu – – – – – – – – – – – – – – His – – – – – – – – Asp 102 Serine Protease and AchE Chymotrypsin Trypsin Elastase Thrombin Plasmin Acetylcholinesterase – Thr – Ile – Asn – Asn – Asp – Ile – Thr – – Tyr – Leu – Asn – Asn – Asp – Ile – Met – – Ser – Lys – Gly – Asn – Asp – Ile – Ala – – Asn – Leu – Asp – Arg – Asp – Ile – Ala – – Phe – Thr – Arg – Lys – Asp – Ile – Ala – – – – – – – – – – – – – – – Asp – – – – – – – Aulani " Biokimia Enzim " Presentasi 1 47 Sigmoidal Curve Effect vo Sigmoidal curve Noncooperative (Hyperbolic) ATP Positive effector (ATP) brings sigmoidal curve back to hyperbolic CTP Cooperative (Sigmoidal) Negative effector (CTP) keeps vo Exaggeration of sigmoidal curve yields a drastic zigzag line that shows the On/Off point clearly Consequently, Allosteric enzyme can sense the concentration of the environment and adjust its activity Off [Substrate] On Aulani " Biokimia Enzim " Presentasi 1 48 Mechanism and Example of Allosteric Effect Kinetics R = Relax (active) Models Cooperation Allosteric site Homotropic R vo (+) S S R S [S] R Concerted Allosteric site A (+) vo S (+) Heterotropic T (+) S Sequential R X [S] T T = Tense (inactive) (+) I vo (-) X (-) T [S] Aulani " Biokimia Enzim " Presentasi 1 T X Heterotropic (-) Concerted 49 Activity Regulation of Glycogen Phosphorylase Covalent modification P A P GP phosphatase 1 AAP T spontaneously AMP Non-covalent ATP Glc-6-P Glucose Caffeine A Glucose Caffeine A P P P A P R P R A P T GP kinase A P A P A P A Aulani " Biokimia Enzim " Presentasi 1 50 Aulani " Biokimia Enzim " Presentasi 1 51