* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Biofilms - Welcome to Cherokee High School
Trimeric autotransporter adhesin wikipedia , lookup
Antibiotics wikipedia , lookup
Human microbiota wikipedia , lookup
Magnetotactic bacteria wikipedia , lookup
Quorum sensing wikipedia , lookup
Marine microorganism wikipedia , lookup
Horizontal gene transfer wikipedia , lookup
Triclocarban wikipedia , lookup
Bacterial taxonomy wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Biofilms
Microbial Biochemistry
Definition of a Biofilm
Biofilms are communities of microorganisms in
a matrix that joins them together and to
living or inert substrates
Biofilms are surface-attached communities of
bacteria, encased in an extracellular matrix
of secreted proteins, carbohydrates, and/or
DNA, that assume phenotypes distinct from
those of planktonic cells
Formation of biofilms in nature
Biofilms offer their
member cells several
benefits
Biofilms are diverse
from their formation
on teeth as plaques
and submerged rocks
in a stream
Biofilms
BIOFILMS may form:
On solid substratums in contact with
moisture
On soft tissue surfaces in living
organisms
At liquid-air interfaces.
Study of biofilms
Formation of multicellular communities
depends on the production of
extracellular substances( matrix)
Diversity in the formation of the matrix
Pathogens that have been
studied for the formation of
biofilms
Staphylococcus aureus
Staphylococcus mutans
Salmonella typhi
Enterococcus faecalis
Pseudomonas aeruginosa
Biofilms – Now a Universal
Feature
Now scientists believe biofilm formation
is a universal feature of all bacteria
Biofilm characteristics
Submerged biofilms seems to form columns
and mushroom like projections that are
separated by water-filled channels
Floating biofilms form a skin or pellicle at the
air- liquid interface – shows organization of
cells with the matrix at the outside
Films that form on the surface of solid media
such as agar or other surfaces
Steps in biofilm formation
Initiation of biofilm formation –
interaction of cells with a surface or
with each other
Films aggregate
Then the cells form an extracellular
matrix
Structure of biofilms are dramatically
different due to the specific organisms
in the film and environmental conditions
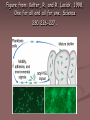
Figure from: Kolter, R. and R. Losick. 1998.
One for all and all for one. Science
280:226-227.
Steps in Biofilm Formation
Exopolysaccharides
Exopolysaccharides
In the glycoclyx
contribute to biofilm
formation
Matrix
Key components of
the matrix are
extracellular
polysaccharides and
proteins
Dead cells have also
been identified in
biofilms
Extracellular DNA is
also important
Biofilm formation
Matrix constituents
Polysaccharides
Carbohydrates significantly impact
bacterial virulence
Bacteria have capsular polysaccharides
and exopolysaccharides
The polysaccharides are not soluble and
do not disassociate with the bacterial
cells
Polysaccahrides
Many bacteria have been found to produce cellulose
This is a novel finding in the case of Salmonella
typhimurium and E. coli
The bacterium Gluconacetobacter xylinus has been
recognized as a cellulose producer
Many other bacteria have genes homologous to the
bcs, bacterial cellulose synthesis genes
Vibrio cholera does not appear to have a gene which
encodes a cellulose – but the bacterium has two
domains homologous to Gluconeacetobacter.
Biofilm Genetics - initiation
Staphylococcal polysaccharide intercellular
adhesin( PIA)
PIA – poly-N-acetyl glucosamine (PNAG)
polymer
PIA polymers present in E. coli
The pga gene is involved with the formation
of biofilms
This gene is similar to the staphylococcal gene
ica
Now Yersinia pestis has been shown to make a
gene homologous to this
Bap Protein
The biofilm associated protein
Structurally similiar to the surface
proteins
Esp of Enterococcus faecalis
mus20 of Pseudomonas aeruginosa
sty2875 of Salmonella typhi
Pseudomonas aeruginosa
Pseudomonas strains were thought to produce
alginate
However alg mutants did not produce changes
in the biofilm formation
Two new polymers have been found in
Pseudomonas polymers
pelA-G gene produces a glucose rich polymer
pslA-O genes produce a mannose rich polymer
Population density and
polysaccharide production
The connection between quorum sensing
and biofilm architecture
Biofilm thickness seems to affect
communication
Quorum sensing mechanism is not
clearly defined but seems to be
essential in the formation of the film
and its channels
Vibrio cholera
Major extracellular polysaccharide in
the biofilm are VPS and the genes, vps
VPS is negatively regulated by hapR.
Hap R mutants produce rugose colonies
and narrow channels in the biofilm
architecture
hapR gene encodes a transcription
factor
hapR is repressed by LuxO
Conjugative pili
Conjugative pili greatly accelerates
initial adhesion and biofilm development
by E. coli
Gram negative bacteria have adhesins at
the tip of its fimbriae
E. coli responds to levels of nutrients
and osmolarity
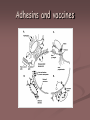
Adhesins
Adhesins are
molecules that are
attached to
bacterial fimbriae
Staphylococcus aureus
E. Coli Adhesins
Adhesins
Bacterium
Adhesins
Receptor
Attachment
Disease
Vibrio cholera
Npili
methylphenylalanine
Fucose and
Mannose
Intestinal
epithelium
Cholera
Bordetella
pertussis
Fimbriae ("filamentous
hemagglutinin")
Galactose on sulfated
glycolipids
pili
Respiratory
epithelium
Whooping cough
Streptococcus
pyogenes
Protein F
amino
terminus of
fibronectin
Pharyngeal
epithelium
Sore throat
E. coli
Type 1 Fimbriae
Species
specific
carboydrate
s
Intestinal
epithelium
Diarrhea
Adhesins and vaccines
Bacterial Cell Characteristics
The phenotypes of the cells include:
a slower growth rate
increased antibiotic resistance
elevated frequency of lateral gene
transfer
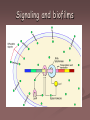
Quorum sensing
Is controlled by at least two different
quorum sensing signals
Acyl-homoserine lactone CAI-1( cholera
autoinducer 1) appears to play a significant
role in biofilm formation
Under low cell density CAI levels are lose
enough to permit LuxO mediated repression
of hapR resulting in VPS production
This bacterium appears to initiate production
of an extracellular matrix under conditions of
low population density presumably before the
establishment of a multicellular community
Signaling in biofilms
Signaling and biofilms
Gram Negative
Gram Positive
Antibiotic Resistance
In the center of the biofilm the
bacteria have greater antibiotic
resistance
Opsonisation of antibiotics
To resistance to phagocytosis
E. faecalis
E. faecalis biofilms on dental root
canals, urethral catheters, uretheral
stents ,and heart valves have been
observed.
While it is not clear that the ability of
E. faecalis to form biofilms is essential
for virulence, it appears that a majority
of clinical isolates do possess the ability
to form a biofilm in vitro
Dental plaque
Genome – genome interactions:bacterial
communities in initial dental plaque –
Paul Kolenbrander et al – Trends in
Microbiology. January 2005.
Found on the enamel of teeth
Epithelial cells of the oral mucosa
Participate in coaggregation which
occurs between different species of
bacteria
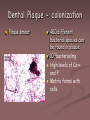
Dental Plaque - colonization
Plaque smear
400 different
bacterial species can
be found in plaque
1010bacteria/mg
High levels of Ca++
and P
Matrix forms with
cells
Definition
Supra gingival –
above gums
Subgingival –below
the gums
Also related to the
tooth surface
Primary Colonizers
The pellicle-coated tooth surface is colonized
by Gram-positive bacteria such as
Streptococcus sanguis, Streptococcus
mutans, and Actinomyces viscosus
These organisms are examples of the
"primary colonizers" of dental plaque.
Bacterial surface molecules interact with
components of the dental pellicle to enable
the bacteria to attach or adhere to the
pellicle-coated tooth surface.
Primary colonization
For example, specific protein molecules found as part
of the bacterial fimbria (hair-like protein extensions
from the bacterial cell surface) on both
Streptococcus sanguis and Actinomyces viscosus
interact with specific proteins of the pellicle (the
proline-rich proteins) with a "lock and key" mechanism
This results in the bacteria firmly sticking to the
pellicle-coating on the tooth surface (Mergenhagen et
al. 1987). Within a short time after cleaning a tooth,
these Gram-positive species may be found on the
tooth surface.
Mechanisms for plaque
formation
two distinct mechanisms: 1 ) the
multiplication of bacteria already
attached to the tooth surface, and 2)
the subsequent attachment and
multiplication of new bacterial species
to cells of bacteria already present in
the plaque mass.
Mechanisms
Two distinct
mechanisms:
1 ) the multiplication of
bacteria already
attached to the tooth
surface
2) the subsequent
attachment and
multiplication of new
bacterial species to
cells of bacteria already
present in the plaque
mass.
Complexity increases
The secondary
colonizers include
Gram-negative
species such as
Fusobacterium
nucleatum,
Prevotella
intermedia, and
Capnocytophaga
species.
Tertiary colonizers
After one week of plaque accumulation,
other Gram-negative species may also
be present in plaque. These species
represent what is considered to be the
"tertiary colonizers", and include
Porphyromonas gingivalis, Campylobacter
rectus, Eikenella corrodens,
Actinobacillus actinomycetemcomitans,
and the oral spirochetes (Treponema
species).
Characteristics
The structural characteristics of dental plaque in this
time period reveal complex patterns of bacterial cells
of cocci, rods, fusiform, filaments, and spirochetes.
In particular, specific associations of different
bacterial forms have been observed. For example, the
adherence of cocci to filaments results in a typical
form referred to as "test-tube brushes" or "corncob" arrays
these structures can be seen in The structural
interactions of the bacteria probably are a reflection
of the complex metabolic interactions that are known
to occur between different plaque microorganisms.
SELECTED BACTERIAL SPECIES FOUND IN DENTAL PLAQUE
Facultative
Gram-Positive
Gram-negative
Spirochetes
Anaerobic
Streptococcus mutans
Streptococcus sanguis
Actinomyces viscosus
Actinobacillus
actinomycetemcomitans
Capnocytophypa species
Eikenella corrodens
Porphyromonas gingivalis
Fusobacterium nucleatum
Prevotella intermedia
Bacteroides forsythus
Campylobacter rectus
Treponema denticola
(Other Treponema species)
Coaggregation
Express components that mediate cell
to cell binding
One cell type in a coaggregation
partnerships, one cell type expresses a
heat-inactivated protease sensitive
surface adhesion
The other partner expresses a
complementary heat stable protein
Biofilm Biochemistry
The production of succinic acid from Campylobacter
species that is known to be used as a growth factor
by Porphyromonas gingivalis. Streptococcus and
Actinomyces species produce formate, which may
then be used by Campylobacter species.
Fusobacterium species produce both thiamine and
isobutyrate that may be used by spirochetes to
support their growth. The metabolic and structural
interactions between different plaque microorganisms
are a reflection of the incredible complexity of this
ecological niche.
Genes and Biofilms
November 2005
Biologist Alejandro Toledo Arana has
identified two genes that regulate the
formation of biofilms in Staphylococcus
aureus
Chronic Infections
The study has been boosted on
discovering their relation to chronic
infections associated to medical
implants
These include those tissues involving
infections of the middle ear, of the
prostate gland, pneumonia in patients
with cystic fibrosis, osteomyelitis
Antibiotic resistance in biofilms
In patients such as those that suffer from
Cystic fibrosis and infections of Pseudomonas,
there is a weighted response in the CD4 T
helper cells( Th1/Th2)
Th1 might improve prognosis for elimination
and management of infections because they
are associated with an influx of phagocytes
and the ingestion of sessile bacteria.
They also ingest biofilm fragments
Survival Strategies of Infectious Biofilms
C.A. Fux, J. W.Costerton et al
Trends in Microbiology
There is a growing concern for antibiotic
resistance in bacteria growing in surfaceadherent biofilms
Many antibiotic assays for susceptibility and
resistance are based upon planktonic or free
cells rather than attached
Chronic and device related infections go
unresolved even when the organisms do indeed
test for antibiotic sensitivity
Biofilm characteristics
Top to bottom gradient of decreasing
antibiotic susceptibility
The gradient originates in the surface layers
of the biofilms where there is complete
consumption of oxygen and glucose
There are patches of antibiotic resistance at
the surface
Proximity of cells lead to horizontal transfer
of genes for resistance
Biofilms and Antibiotics
The diffusion of antibiotics in biofilms
has been studied
Beta lactamase producing bacteria
increase enzyme production in response
to antibiotic treatment
The enzyme accumulates in the matrix
of the biofilm thereby inactivating the
antibiotic
Mar Operon
Multiple Antibiotic Resistance operon (Mar)
is chromosomal, and encodes for permease
proteins (AcrB) which actively export a wide
range of xenobiotics from bacterial cells.
Mar is widely distributed. Recent reports
show that Mar can be regulated not only by
exposure to sub-MIC levels of antibiotic, but
also through slow growth rate, the stringent
response and a number of other unrelated
stimuli.
Mar II
It is not regulated through homoserinelactone
but does appear to be part of a global
regulatory system that also controls
exopolymer biosynthesis.
This operon is of major interesdt since it is
likely that this would be switched on in
biofilms and might be a major factor in the
high level antibiotic resistance observed in
biofilms.
Endocarditis
Biofilm of bacteria +
host components on
valve = vegetation
Requires prior valve
injury
200X increase in
antibiotic resistance
Rabbit model: block
biofilm formation -->
acute virulent infection
Infectious Kidney Stones
15-20% involve urinary
tract infection
Bacterium --> biofilm -->
mineralization
Causative organisms
have urease
urea --> NH4 + H2CO3
Biofilm concentrates
urease --> crystal
formation
CF
Mutation in chloride
channel in epithelial
cells
1st stage: intermittent
infections
2nd stage: permanent
infection with
Pseudomonas aeruginosa
Mucoid type overproduce alginate
Antibiotic resistance
Microarrays
Used to assess the
genes present in
different stages of
biofilm formation
In Staphylococcal
biofilms the same
genes are active the
sar A staphylococcal
accessory regulator
and the ica ADBC
regulator
Microarrays
One of the best
ways to evaluate
gene expression
DNA chips are used
for a solid support
These are made of
silicon or glass
They have DNA
attached in orderly
arrays
Microarrays( 1)
The DNA is deliverd to specific position
on the chip using tiny pins to apply a
solution
The spots are treated and dried in
order to bind the DNA
Usually cDNA
cDNA is prepared from mRNA
These pieces of DNA are usually 5005000 nucleotides long
Commercial chips( II)
Oligonucleotides about 25 bases in length can
be synthesized and placed directly on the chip
The chip is 1.3 cm on a side and can have over
200,000 addressable positions
The probes are often expressed sequence
tags( ESTs)
The nucleic acids to be analyzed are isolated
and labeled with fluorescent reporter groups
The DNA or target nucleotides are incubated
with the fluorescent groups and then washed
The chip is scanned with lasers
Preparation of Microarrays
Interpretation of microarrays
Agents for the destruction of
biofilms( Industrial biocides)
(alexidine, chlorhexidine,
polyhexamethylene biguanides),
monophenylethers (phenoxyethanol) and
quaternary amonium compounds
(cetrimide, benzalkoniums) and have
demonstrated biochemical bases for the
activities and associated mammalian cell
toxicities of thiol interactive agents
(bronopol, isothiazolones).