* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Running GBrowse and DAS/1 on GUS
Oracle Database wikipedia , lookup
Concurrency control wikipedia , lookup
Microsoft SQL Server wikipedia , lookup
Extensible Storage Engine wikipedia , lookup
Entity–attribute–value model wikipedia , lookup
Microsoft Jet Database Engine wikipedia , lookup
Open Database Connectivity wikipedia , lookup
Relational model wikipedia , lookup
Functional Database Model wikipedia , lookup
GBrowse-related work at ApiDB
Haiming Wang
ApiDB Bioinformatics Resource Center
Dr. Jessica Kissinger Group
GMOD Nov 2007
•
•
•
Same database schema – GUS
Multi-species – comparative analysis
Graphical for protein data
Database adaptor vs GFF
Pros:
• Live interactive with database
• Flexible with database schema – GUS, Chado
• Support complex data type, e.g. synteny, protein data
Cons:
• Relatively slow – database tuning + bulkSubfeatures
• Familiar with SQL
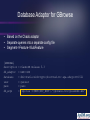
Database Adaptor for GBrowse
• Based on the Chado adaptor
• Separate queries into a separate config file
• Segment->Feature->SubFeature
[GENERAL]
description
db_adaptor
database
user
pass
db_args
=
=
=
=
=
=
PlasmoDB Release 5.3
DAS::GUS
dbi:Oracle:sid=crypto;host=rad.rcc.uga.edu;port=1521
gususer
pass
-sqlfile =>$ENV{DOC_ROOT}.'/gbrowse.conf/plasmodb.xml'
features
segment
PBrowse
Synteny Browser - SynView
Steve Fisher will present it in details later
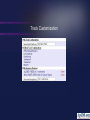
Track Customization
init_code = sub hover {
use HTML::Template;
my ($name, $data) = @_;
my $tmpl = HTML::Template=>new(filename=>$ENV{ROOT}.'/gbrowse/hover.tmpl');
$tmpl->param(DATA => [ map {{@$_ > 1 ? (KEY=>$_->[0],
VALUE => $_->[1]):(SINGLE => $_->[0])}} @$data]);
my $str = $tmpl->output;
$str =~ s/'/\\'/g;
return qq{" onMouseOver="$cmd;return escape('$str')};
}
link_target
= sub { my $f
my $name
my $start
my $stop
my ($freq)
my ($tag)
my @data;
push @data, [
push @data, [
push @data, [
push @data, [
return hover(
}
=
=
=
=
=
=
shift;
$f->name;
$f->start;
$f->stop;
$f->get_tag_values('RawCount');
$f->get_tag_values('Sequence');
'Location:' => "$start..$stop" ];
'Sequence:' => $tag ];
'Library:'
=> $lib ];
'Frequency:' => $freq];
"Sage Tag - Temp ID $name", \@data);
Future Works
•
•
•
•
•
•
Performance tuning – SQL, bulksubfeatures, AJAX tooltips
Unification with web inferface WDK
All projects share same gbrowse configuration
Be able to divide the .conf and .xml files into sub files
Be able to include/exclude based on project
Re-using sql and perl across tracks, e.g. duplicate code