* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Poster at European Conference on Computational Biology 2003
Concurrency control wikipedia , lookup
Data vault modeling wikipedia , lookup
Business intelligence wikipedia , lookup
Versant Object Database wikipedia , lookup
Information privacy law wikipedia , lookup
Open data in the United Kingdom wikipedia , lookup
Relational model wikipedia , lookup
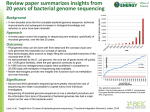
MolliGen, a database dedicated to the comparative genomics of Mollicutes Aurélien Barré, 2Pascal Sirand-Pugnet, 2Xavier Foissac, 3Eduardo P. C. Rocha, 1Antoine de Daruvar and 2Alain Blanchard 1 1Centre de Bioinformatique Bordeaux (CBiB), Université Victor Segalen Bordeaux 2; 2UMR GDPP, INRA-Université Victor Segalen Bordeaux 2 3CNRS URA 2171, Institut Pasteur, Paris-FRANCE Bacteria belonging to the class Mollicutes were among the first ones to be selected for complete genome sequencing because of the minimal size of their genome and of their pathogenicity for humans and a broad range of animals and plants (1,2,3) (Figure 1). Comparative genomics analysis is difficult to carry out without a suitable platform gathering not only the original annotations but also relevant information available in public databases or obtained applying common bioinformatics methods. With the aim of solving these difficulties, we have developed a web-accessible database named MolliGen. Original genome data (GenBank) External public resources (NCBI COG, KEGG, ) HTML report input / output files Figure 1 : Mollicute phylogenic tree. In green genomes integrated in MolliGen,in red others available complete genomes 1st data level (core) -mapping genes on metabolic pathway Structure 2nd data level -building orthology relationship …. Information, extracted from various databases or computed locally, are stored as structured data in MolliGen relational database, which consists of two levels : Export manager Query, Browse, Visualise individual genomes Browse, Visualise, Align multiple genomes (comparative genomics) 1. the first one comprising most basic information and stored as the database core 2. the second containing all other computed information (domains, metabolic pathway, homology, …) Such a structure (Figure 2) allows to easily add new organisms (by extending the core) or new information (by extending the second level). Figure 2 : MolliGen schema for data integration and accession via the web Query MolliGen provides access to integrated data through a web form in which query is dynamically built by the user (Figure 3A). Results can be obtained either for only one species or globally, with links to other information and bioinformatic methods (Figure 3B-D). Comparison • A multi-genomes browser developed for MolliGen allows to visualize more than one genome and to display relationships between them. (Figure 3D) • A clickable dot-plot representation allows to visualise relationships between two genomes over their full length. • A metabolic pathway viewer, based on KEGG predictions (4) for enzymatic functions, has been developed to show graphically resemblance between set of genome(s) for a selected pathway. • A multi proteome differential queries can be performed to find genes specific for a genome (or a set of genomes) having homologs in a targeted group of other genomes. (Figure 1E-F). A third group of genomes can be selected as an exclusion genome set where no homologs must be found. Conclusion MolliGen centralizes and integrates heterogeneous information about mollicutes in a database. New genomes sequences and information will Figure 3 : MolliGen interface overview be added as they will become publicly available. This database will also be used as an aid for the re-annotation of these genomes, using 1.- Frey, J. (2002) Animal mycoplasmas. In Herrmann, R. (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, pp. 73-90. 2. - Blanchard, A. and Bébéar, C.M. (2002) Human mycoplasmas. In Herrmann, R. (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, pp. 45-71. 3. - Bove, J.M., Renaudin, J., Saillard, C., Foissac, X. and Garnier, M. (2003) SPIROPLASMA CITRI, A PLANT PATHOGENIC MOLLICUTE: Relationships with Its Two Hosts, the Plant and the Leafhopper Vector. Annu Rev Phytopathol, 41, 483-500. 4. Kanehisa M, Goto S. (2000 ) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 2730. homology relationship between them. MolliGen is publicly available at http://cbi.labri.fr/outils/molligen/.