* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Sudarshan - CSE, IIT Bombay
Survey
Document related concepts
Transcript
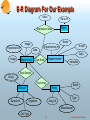
Data Representation in Bioinformatics S. Sudarshan Computer Science and Engg. Dept. I.I.T. Bombay Data Representation Goal: Represent data in an intuitive and convenient manner Without unnecessary replication of information Making it easy to write queries to find required information Supporting efficient retrieval of required information Data Models Ad-hoc file formats (not really data models!) XML (Extensible Markup Language) Relational data model Entity-relationship (ER) data model Object-relational data model Object-oriented data model 2 S. Sudarshan, IIT Bombay Data Representation in Genomics Most common approach: Text Files E.g. GenBank: GenBank Example Advantage: Easy to export data to others (integrating datasets is not my problem!) Drawback: Makes it hard to integrate information from different sources This is essential for many applications e.g. comparative studies Multiplicity of formats makes interoperation difficult Reading a particular file format requires a program designed to “parse” that file format No standard query language Complex queries needed to integrate data from different sources Several efforts to create standard file formats are based on a “tag” language called XML 3 S. Sudarshan, IIT Bombay Genbank Example LOCUS DEFINITION ACCESSION VERSION KEYWORDS SOURCE ORGANISM REFERENCE AUTHORS TITLE JOURNAL FEATURES source AB020037 300 bp mRNA EST 11-MAY-1999 AB020037 Phaseolus vulgaris library (Watanabe T) cDNA, mRNA sequence. AB020037 AB020037.1 GI:4783241 EST. Phaseolus vulgaris. Phaseolus vulgaris Eukaryota; Viridiplantae; Streptophyta; Embryophyta; … 1 (bases 1 to 300) Watanabe,T., Watanabe,T, …. Partial cDNA G.max calnexin homologue from P.vulgaris Unpublished (1999) Location/Qualifiers 1..300 /organism="Phaseolus vulgaris" /db_xref="taxon:3885" /clone_lib="Phaseolus vulgaris library (Watanabe T)" 92 a 50 c 82 g 76 t BASE COUNT ORIGIN 1 gacctgcgat cttctacgaa tcattcgatg aggattttca agatcgttgg atcgtgtctc 61 agaaagagga atacagtggt gtctggaaac atgccaagag tgagggacat gatgatcatg 121 gtcttcttgt cagtgagaaa gcaagaaaat atgccatagt gaaggaactt gacaaggcag 181 tgagtctcag ggatggaact gttgttctcc agtttgaaac tcggcttcag aatggacttg 241 aatgtgaagg agcatatata aaatatctcc gaccacaggg atgctggatg ggaactctaa // 4 S. Sudarshan, IIT Bombay XML: Extensible Markup Language Simple XML example E.g. <faculty> <faculty-member facid=12349> <name> S.Sudarshan </name> <email> [email protected]</email> </faculty-member> <faculty-member facid=12987> <name> Pramod Wangikar</name> <email> [email protected]</email> </faculty-member> </faculty> Each piece of text enclosed by matching tags <xyz> … </xyz> is called an element Elements may have attributes (such as facid in the example above) DTD (Document Type Descriptor) specifies allowed element, attributes of each element, and what elements may appear within each element (and how many times and in what order). Each application defines a standard set of elements (including how they are nested) and attributes for each element 5 S. Sudarshan, IIT Bombay XML Representation (Cont.) Ad-hoc file representations are being replaced by standard XML representations (see e.g. http://i3c.open-bio.org) Examples: Gene Expression Markup Language (GEML) (http://www.geml.org) – (GEML 2.0 white paper: http://www.geml.org/docs/GEML2_0.pdf) Bioinformatic Sequence Markup Language (BSML) (http://www.labbook.com/products/xmlbsml.asp), and many others – Earlier GenBank example in in XML (BSML) Benefits Standardization will simplify inter-operation and data sharing XML tagged datasets are easy to read and comprehend Parsing of datasets is simple with XML Problems: Standards take time to develop (for human/political reasons) More than one standard may evolve People may not adopt standards, sticking to old formats Support for querying on XML data is still poor (but will improve) 6 S. Sudarshan, IIT Bombay Genbank Example in XML (BSML) <?xml version="1.0" ?> <records> <record> <locus name="AB020037" bp="300" strands="" molecule="mRNA" geometry="linear" division="EST" date="11-MAY-1999"/> <definition> <![CDATA[ AB020037 Phaseolus vulgaris library (Watanabe T) Phaseolus vulgaris cDNA, mRNA sequence ]]> </definition> <accession name="AB020037"/> <version accession="AB020037.1" gi="4783241"/> <keywords> EST </keywords> …….. ……. 7 S. Sudarshan, IIT Bombay Present vs. Future XML databases are coming but not quite here yet In alpha versions at best Some relational database provide support for storing XML data, but no support or poor support for quering complex XML data XML query language is still being standardized (XQuery) Initial XML query implementations likely to be poor compared to relational query implementations which are mature Interesting query execution/optimization problems to be solved, even ignoring bioinformatics Relational data can be viewed as a special case of XML data Issues we describe in next few slides also applicable to XML representation XML good for data exchange Can easily convert simple XML data to relations Perhaps a few years down the road we can use XML for querying genomics data 8 S. Sudarshan, IIT Bombay What are Relations Attributes or columns Name E-mail Department Pramod Seshadri Uday Sudarshan [email protected] [email protected] [email protected] [email protected] Chem. Engg. Mech. Engg. Elec. Engg. Comp. Sci. Tuples or rows faculty 9 S. Sudarshan, IIT Bombay Relational Representation The relational data model is widely used and supported by all the popular commercial database systems Allows 1) information to be broken up into logical units, and then 2) recombined in different ways as required Great for queries involving information from multiple original sources Can easily gather related information e.g. information about a particular gene from multiple datasets/experiments Entity Relationship (E-R) Model: Higher level model than the relational model Often used for design, and then converted (automatically or manually) into a relational schema Has several diagrammatical representations Widely used 10 S. Sudarshan, IIT Bombay Entities and Relationships A database can be modeled as: a collection of entities, relationship among entities. An entity is an object that exists and is distinguishable from other objects. Example: gene, protein, experiment, organism, person Entities have attributes An entity set is a set of entities of the same type that share the same properties. Example: set of all persons, companies, trees, holidays Relationships provide connections between two or more entities E.g. Which genes were involved in which experiment 11 S. Sudarshan, IIT Bombay Example ER Diagram for Microarray Data Entities represented by boxes, (binary) relationships by lines with names and optional attributes See www.bioinf.man.ac.uk for a more realistic version (the MaxD database) Gene gene-id sequence …… Expt-Exptr Expression-value value Expt-Sample Experiment Experiment-Id Date Image Notation * Many-to-one 1 Expt-Array 12 Experimenter Experimenter-Id Name E-mail Dept. Institution Sample Sample-Id Organism Cell-type {Drug-Ids} Array Array-Id Manufacturer Type Batch S. Sudarshan, IIT Bombay Schema Diagrams for MicroArray Data Schema diagrams show multiple relations and their interconnections Lines link foreign key with referenced relation Experimenter Experimenter-Id Name E-mail Dept. Institution Experiment Experiment-Id Date Experimenter-Id Sample-Id Array-Id Image Expression-Value Experiment-Id Gene-Id value Sample Sample-Id Organism Cell-type {Drug-Ids} Multivalued attribute Array Array-Id Manufacturer Type Batch Gene Gene-Id sequence 13 S. Sudarshan, IIT Bombay Modeling Protein Data (from Paton & Goble) 14 S. Sudarshan, IIT Bombay Schema Diagrams vs. ER Notation Don’t confuse ER diagrams with schema diagrams Differences: In ER diagrams: lines have names There are no explicit foreign key attributes In schema diagrams Lines don’t have names, but represent foreign key relationships Foreign key attributes must be explicitly represented Relationships in ER diagrams get converted to separate relations and/or foreign key relationships (more on this later) 15 S. Sudarshan, IIT Bombay Query Languages Language in which user requests information from the database. Categories of languages Procedural E.g. C/C++/Java Advantage: Powerful, can specify any query by programming Disadvantage: Interfacing directly to database is cumbersome non-procedural Web forms! SQL Advantage: – Can specify query “declaratively” and let database system figure out best way of finding answers – Supports queries of medium complexity Specialized languages More complex queries (e.g. data mining such as classification and clustering) implemented in procedural language, with SQL acting as interface to database 16 S. Sudarshan, IIT Bombay Problems of Diversity Many different databases Multiple databases for each of genome, proteome, transcriptome, metabolome (and perhaps any other *ome you choose to add!) Need to cross-reference between these databases Need an ontology to ensure consistent and unique names Instability Names, data, even models keep changing Modeling secondary information Annotations, typically text based 17 S. Sudarshan, IIT Bombay Problems in Querying Querying What query languages to use? (AceDB (SGD), Icarus (SRS), SQL?) OO API (Corba based interfaces proposed by OMG/EMBL) Querying and text mining on annotations Queries that combine multiple databases and paradigms E.g. genome, proteome and annotations (text data) Browsing and visualization Generate hyperlinks in data automatically for browsing Visualization for sequence data, protein structures, to depict correlations, etc 18 S. Sudarshan, IIT Bombay Problems of Scale and Distribution Problems of scale Genome: hundreds of gigabytes to terabytes (1012 bytes) Transcriptome (Microarray): Each chip has 10,000 measurements + image Millions of experiments – on different species/individuals/cells/conditions … – Total: 1 petabyte/annum (1015 bytes) Bottom line: too big to hold everything locally Ideally: provide integrated view of all data, and fetch actual data on demand Limited access patterns Can usually access data only via predefined Web forms 19 S. Sudarshan, IIT Bombay Problems of Database Representation Efficiency and flexibility of use are often at odds E.g. the Expression-Value table in our schema can be huge Array representation may be better but less convenient for users Alternative: use one attribute for each gene – no database efficiently supports relations with thousands of attributes – But this is natural to lay users Similarly: user may want one relation for each of millions of experiments Ideal: flexible view combined with efficient implementation underneath, plus query languages that offer metadata capabilities E.g. “for all relations whose name is in table N” 20 S. Sudarshan, IIT Bombay References Online information Heaps and heaps of sites, many with actual data freely available data may be worth what you paid for it! Tutorial on Information Management for Genome Level Bioinformatics, Paton and Goble, at VLDB 2001: http://www.dia.uniroma3.it/~vldbproc/#tut European Molecular Biology Network http://www.embnet.org/ Univ. Manchester site (with relational version of Microarray data representation, and links to other sites) http://www.bioinf.man.ac.uk Database textbook with absolutely no bioinformatics coverage (shameless sales pitch ): Database System Concepts 4th Ed by Silberschatz, Korth and Sudarshan (should come out in Indian edition in a few months) 21 S. Sudarshan, IIT Bombay End of Talk Relational Schema Design Problems Many flat file formats have lots of columns: E.g. Drug-effect Drug1 Drug2 Drug3 … Drug-n Cancer1 Cancer2 Cancer3 …. Cancer-m Beware: Such structures are nice for humans to read (are called crosstabs), BUT Most databases cannot support relations with many columns! And querying data with such columns is more complicated Solution: use a schema drug-effect(cancer-type, drug, effect) Alternative solution: use arrays to represent some such information (supported by some databases) 23 S. Sudarshan, IIT Bombay Relational Schema Design Problems (Cont.) Another common mistake: having many relations with same attributes E.g. one relation for each cancer type, or one relation for each drug Cancer1(…), Cancer2(…), …, Cancer-n(…) Most databases can handle only hundreds or a few thousand relations efficiently Querying becomes more complicated when there are many relations Solution: Replace many relations with same attributes by a single relation with the same attributes, plus an extra attribute storing the name Cancer(Type, …) 24 S. Sudarshan, IIT Bombay Alternative E-R Notations Crow’s feet notation: Total participation (each entity participates in at least one relationship) is indicated by an extra bar 25 R1 R2 S. Sudarshan, IIT Bombay E-R Diagram For Our Example Value Gene-Id Expression-Value Gene Name Image Experiment-Id E-mail Experimenter-Id Date Dept. Image Experiment Experimenter Expt-Exptr Institution Expt-Sample Drugs Expt-Array Sample-Id Batch Array Sample Array-Id Organism Type Manufacturer Cell Type 26 S. Sudarshan, IIT Bombay Relational Schema Design Principles Redundancy E.g. Array-genes(.., fragment-seq, gene-seq, gene-mutations, …) is better represented as – Array-genes( fragment-seq, gene-id) – Gene(gene-id, gene-seq, gene-mutations) Otherwise data is replicated unnecessarity – I.e. mutation information is stored multiple times Redundancy can be useful for better query performance, but should be used in a thought-out manner, not by accident Inability to express information E.g. if a gene is not stored in Array-genes we cannot store its mutation information 27 S. Sudarshan, IIT Bombay Basic SQL Queries Find the image for experiment number 1345 select image from experiment where experiment-id = 1345 Find the experiment-id and image of all experiments involving e-coli select experiment-id, image from experiment, sample where experiment.sample-id = sample.sample-id and sample.organism = ‘e-coli’ All combinations of rows from the relations in the from clause are considered, and those that satisfy the where conditions are output 28 S. Sudarshan, IIT Bombay Complex Queries and Views A view consisting of experiments with number of active genes create view expt-active-genes as select experiment-id, count (gene-id) as active-cnt from experiment, expression-value where expression-value.experiment-Id = experiment.experiment-Id and value > 2 group by branch-name Find number of active genes in experiment E-123 select active-cnt from expt-active-genes where expirement-Id = ‘E-123’ 29 S. Sudarshan, IIT Bombay