* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Nitrogen Cycle
Horizontal gene transfer wikipedia , lookup
Quorum sensing wikipedia , lookup
Microorganism wikipedia , lookup
Hospital-acquired infection wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Human microbiota wikipedia , lookup
Phospholipid-derived fatty acids wikipedia , lookup
Triclocarban wikipedia , lookup
Disinfectant wikipedia , lookup
Anaerobic infection wikipedia , lookup
Marine microorganism wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Magnetotactic bacteria wikipedia , lookup
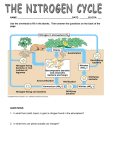
Nitrogen Cycle • Global Nitrogen Budget • Nitrogen Transformations • Denitrifying Bacteria • Nitrifying Bacteria • Nitrogen Fixation • Rhizobium-Legume Symbiosis Nitrogen Facts • Nitrogen Oxidation States: – – – – – Ammonia and organic amines = Nitrogen Gas (N2) = Nitrous Oxide (N2O) = Nitrite ion (NO2-) = Nitrate ion (NO3-) = -3 0 +1 +3 +5 • Sources of Nitrogen on a Global Scale: – – – – Agriculture (fertilizers and livestock wastes) Human waste Combustion of fuels Natural N2-fixation (biological and geothermal) Agriculture Inputs 7.1 mmol N m-2 y-1 for each kg N ha-1 y-1. Scale for agriculture is 7 x that for fossil fuel. Fossil Fuel Combustion Inputs Ammonium is also assimilated by plants, bacteria and fungi! Dissimilatory nitrate reduction is performed by anaerobic respiring bacteria. Nitrification • One of two types of chemolithotrophs – Ammonium Oxidizing Bacteria (AOB): form nitrite. – Nitrite Oxidizing Bacteria (NOB): form nitrate • All require oxygen as terminal electron acceptor; mostly autotrophic • Often AOB and NOB found together in aerobic soils and aquatic habitats to collectively oxidize ammonium to nitrate. • Phylum: Proteobacteria (α, β, γ, γ) α-Proteobacteria β-Proteobacteria Nitrosomonadaceae γ-Proteobacteria Nitrococcus Nitrosococcus Why care about nitrifying bacteria? • Loss of N-fertilizers and groundwater contamination. • Most mineral fertilizers are added as ammonium. • Presence of oxygen and neutral pH promotes thriving nitrifying bacterial community. • Soil particles are negative charged! “Likes repel”. • Rain causes leaching. • Oxygen depletion causes denitrification and the anammox process. Anaerobic Ammonium Oxidation NH4+ + NO2- → N2O + H2O +2H+ Phylum: Planctomycetes Denitrification • Type of dissimilatory nitrate reduction where nitrous oxide or nitrogen gas is released as an end product of anaerobic respiration. – Compare with that of E. coli; nitrite end product. – Compare to Clostridium (Firmicutes), and Desulfovibrio (δ-Proteobacteria); ammonium end product. • Found in anaerobic habitats, such as sediments and saturated soils, yet in close proximity to nitrifying bacteria that supply nitrate for their respiration. • Phyla for denitrifying bacteria: – γ-Proteobacteria (Pseudomonas) – Firmicutes (Low G+C Gram Positives) (Bacillus) Sediment or Hypereutrophic Lake Nitrogen Cycling Profiles N2-Fixation • Nitrogenase enzyme complex and 8 ATP needed for reducing N2 to ammonia. • Must have low oxygen level (< 10% atmospheric). • Different strategies for reducing O2 in aerobic habitats. • Extremely diverse phylogenetic distribution in Prokaryotes (never Eukarya). • Example of horizontal gene transfer throughout evolution. Trichodesmium spp. Rhizobium-Legume Symbiosis Bacteria gets protection and organic nutrients; legume gets a supply of nitrogen for growth. 60 % terrestrial nitrogen fixation Chemotaxis to the root epidermis cells which exude polyphenolic flavonoid inducers of Rhizobium nod genes. Legume (pod plants), only release the flavonoids when under nitrogen stressed soil conditions. A result of nod gene induction is expression of rhicadhesin at the outer membrane surface. Root hair surface lectins specifically bind rhicadhesin; Nod factor proteins induce roothair curling. Rhizobium stimulates the formation of an infection thread that the bacterium moves into and grows within. Ultimately infection is within plant root cortex cells, where Rhizobium transforms to a non-motile bacteroid state; covered in host membrane (aka peribacteroid membrane). Transformation continues to form the sybiosome where in the bacterium is now pleomorphic and begins fixing N2. Leghemoglobin controls oxygen supply to bacteroid. Active nodule as indicated by pink. Comparison with other related legume nodulating bacteria