* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Outline for Unknown Bacteria Report
Microorganism wikipedia , lookup
Quorum sensing wikipedia , lookup
Horizontal gene transfer wikipedia , lookup
Phospholipid-derived fatty acids wikipedia , lookup
Bioremediation of radioactive waste wikipedia , lookup
Triclocarban wikipedia , lookup
Disinfectant wikipedia , lookup
Marine microorganism wikipedia , lookup
Human microbiota wikipedia , lookup
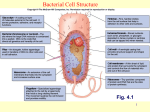
Bacterial cell structure wikipedia , lookup
Outline for Unknown Bacteria Report
David J. Sanchez, Ph.D.
The purpose of this report is the present a thorough analysis of an unknown bacterial
culture and explain to the reader how the unknown’s identity was determined. The
format is in another document contained on this website. This information is on how to
determine the identity of your unknown bacteria.
Differential Staining in Practice
Differential Stains allow one to distinguish a particular type of bacterial species. Use of
these stains is a key step (yet not the entirety) of the process in determining the identity of
an unknown or disease causing bacteria.
Gram + bacteria and Gram – bacteria effectively split the bacterial kingdom into two
separate categories.
Acid Fast + bacterial cells are indicative of one main bacterial species, Mycobacteria.
This is also a Gram + species. Thus no Gram – bacteria is Acid Fast +.
Endospore + bacterials cells are indicative of two potential bacterial species, Bacillus and
Clostridium. All Endospore + bacteria are Gram +.
The following flow chart summarizes the use of these three main differential stains:
1*+2&3&-" &! " #" $%" &4 +*+,5 -"( 6$" &
! "#$%$%&'#%( '! ) #*+'
! " #" $%" &' ( ) *+,-( &
. ,$) +/ 0,+!"# $%& "' (%)*"
# $%& "+"
, %- )..)/"
01- - 12%- )..)"
# $%& "+"
01- - )"
# $%& "3"
, %- )..)"
# $%& "3"
01- - 12%- )..)"
# $%& "3"
01- - )"
7 8 9: ;&7 $&8 *<+,&1*( -" -" =&
. ,$/ 0) +>&? / / -6$" ( @
&A" B$,5 ( 6$""
. ,$) +/ 0,+!"4*51671$8/"
9- )5": %6("' (%)*)*; "
4*51671$8"+"
9- )5": %6("+"
4*51671$8"+"
9- )5": %6("<"
. ,$C@
+5 D&
E+/ $&' $*<&1*( -" >&
' ( ) *+,-( &F +" 0>;&
, #- $../0'
4*51671$8"<"
9- )5": %6("+"
' ( ) *+, -( &F +" 0>;&
1 2- 34#- "+5$/6 '
4*51671$8"<"
9- )5": %6("<"
' ( ) *+,-( &F +"0>;&
7352%+4#- "+5$/6 '
Phenol Red Fermentation Test Results
The first +/- is about the acid (yellow is +, red is -) and the second is about a bubble in
the tubes (bubble is +, no bubble is -). Thus a
Gram Positive Cocci:
Staphylococcus
Gram positive cocci
Catalast positive,
S. aureus
Colony
Yellow
Gelatinase
Positive
Phenol Red Mannitol Yellow
S. epidermidis
White
Negative
Red
Micrococcus
Gram positive cocci
Nonmotile, catalase positive
Colony
Motility
Mannose Use
Lactose Use
Gelatinase Use
Citrate Use
M. agilis
Red
+
+
-
M. luteus
Yellow
+
-
M. roseus
Pink
+
-
M. sphagni
+
+
?
+
+
-
M. thermoresistin
+
+
+
-
Mycobacterium
Mycobacterium
Weak Gram positive cocci/coccobacillia
Catalase positive
Acid Fast
M. smegmatis
Pigment
Nitrate Use
+
Iron Uptake
+
Citrate Use
+
Mannitol Use
+
Arabinose Use
+
Gelatine Use
Starch Use
+
Casene
-
Bacillus
Bacillus
Gram positive, bacilli to coccobacilli, endospore positive
Usually motile and catalase positive
Lactose Use
Sucrose Use
Mannitol
Dextrose Use
Citrate use
B. megaterium
-
B. cereus
+
+
-
B. subtilis
+
-
Enterobacteria are a diverse set of Gram negative bacilli/coccobacilli
These are tricky… follow all the logic patterns present! Use the Phenol Red tests in the
second page if you are confused here.
Gram
Citrate
H2S
Motility
Gelatin
Arabinose
Lactose
Mannitol
Mannose
Sucrose
Nitrate
Catalse
S. flexni | E. Arogenes |S. Sonnei |Citrobacter|E. Coli | K. pneumoniae
+
+
+
Variable
+
+
+
?
+
+
+
+
+
+
?
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
?
?
+
+
+
+
+
+
+
+
+
+
+
+
+