* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Applying Bayesian networks to modeling of cell signaling pathways
Survey
Document related concepts
Transcript
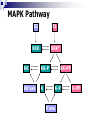
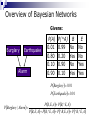
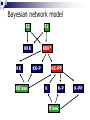
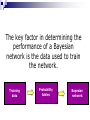
Applying Bayesian networks to modeling of cell signaling pathways Kathryn Armstrong and Reshma Shetty Outline Biological model system (MAPK) Overview of Bayesian networks Design and development Verification Correlation with experimental data Issues Future work MAPK Pathway E2 E1 KKK KKK* KK KK-P KK’ase K KK-PP K-P K’ase K-PP Overview of Bayesian Networks Givens: Burglary Earthquake Alarm P(A) 0.01 0.80 0.10 0.90 P(^A) 0.99 0.20 0.90 0.10 B No Yes No Yes E No No Yes Yes P Burglary 0.01 P Earthquake 0.01 P B, E, A PB,^E, A PBurglary | Alarm PB, E, A P B,^E, A P^B, E, A P ^B,^E, A Bayesian network model E2 E1 KKK KK KK’ase KKK* KK-P KK-PP K K-P K’ase K-PP Simplifying Assumptions Normalized concentrations of all species Discretized continuous concentration curves at 20 states Considered steady-state behavior The key factor in determining the performance of a Bayesian network is the data used to train the network. Training data Probability tables Bayesian network Network training I: Data source Current experimental data sets were not sufficient to provide enough information Relied on ODE model to generate training set (Huang et al.) Captured the essential steady-state behavior of the MAPK signaling pathway Network training II: Poor data variation Network training III: incomplete versus complete data sets 1D x 4 E1 4D E2 MAPKPase MAPKKPase Time = (# samples) x 4 Time = (# samples)4 Verification: P(Kinase | E1, P’ases) Huang et al. Bayesian network Verification: P(E1 | MAPK-PP, P’ases) Correlation with experimental data C.F. Huang and J.E. Ferrell, Proc. Natl. Acad. Sci. USA 93, 10078 (1996). Correlation with experimental data J.E. Ferrell and E.M. Machleder, Science 280, 895 (1998). Where does our Bayesian network fail? Where does our Bayesian network fail? Inference from incomplete data E2 E1 KKK KK KK’ase KKK* KK-P KK-PP K K-P K’ase K-PP Future work Time incorporation to represent signaling dynamics Continuous or more finely discretized sampling and modeling of node values Priors Bayesian posterior Structure learning Open areas of research Should steady state behavior be modeled with a directed acyclic graph? Cyclic networks Theoretically impossible Hard, but doable Need an alternate way to represent feedback loops Why use a Bayesian network? ODE’s require detailed kinetic and mechanistic information on the pathway. Bayesian networks can model pathways well when large amounts of data are available regardless of how well the pathway is understood. Acknowledgments Kevin Murphy Doug Lauffenburger Paul Matsudaira Ali Khademhosseini BE400 students References http://www.cs.berkeley.edu/~murphyk/Bayes/bayes.html http://www.ai.mit.edu/~murphyk/Software/BNT/usage.html A.R. Asthagiri and D.A. Lauffenburger, Biotechnol. Prog. 17, 227 (2001). A.R. Asthagiri, C.M. Nelson, A.F. Horowitz and D.A. Lauffenburger, J. Biol. Chem. 274, 27119 (1999). J.E. Ferrell and R.R. Bhatt, J. Biol. Chem. 272, 19008 (1997). J.E. Ferrell and E.M. Machleder, Science 280, 895 (1998). C.F. Huang and J.E. Ferrell, Proc. Natl. Acad. Sci. USA 93, 10078 (1996). F. V. Jensen. Bayesian Networks and Decision Graphs. Springer: New York, 2001. K.A. Gallo and G.L. Johnson, Nat. Rev. Mol. Cell Biol. 3, 663 (2002). K.P. Murphy, Computing Science and Statistics. (2001). S. Russell and P. Norvig. Artificial Intelligence: A Modern Approach. Prentice Hall: New York, 1995. K Sachs, D. Gifford, T. Jaakkola, P. Sorger and D.A. Lauffenburger, Science STKE 148, 38 (2002). Network training IV: final data set E1 E2 (P’ase) MAPKKPase MAPKPase MAPK-PP 0 0 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 1 1 0 0 1 0 0 0 0 1 0 1 0 0 1 1 0 0 0 1 1 1 0 1 0 0 0 1 1 0 0 1 1 1 0 1 0 1 1 0 1 1 0 1 1 0 0 1 1 1 0 1 0 1 1 1 0 0 1 1 1 1 0 Network training V: Final concentration ranges Network training III: Observation of all input combinations 1D Visualization E1 E2 3D Visualization E2 E1 MAPKPase MAPKKPase MAPKKPase 4D Visualization 2D Visualization Time = (# samples)4