Computational Pharmacology - Carnegie Mellon School of
... Outline Drug Discovery Approach • use the information in the databases and infer information that is not provided directly by genomics and proteomics data: higher level information => piece together all available information - to get detailed picture of a molecular process (or disease) - to identif ...
... Outline Drug Discovery Approach • use the information in the databases and infer information that is not provided directly by genomics and proteomics data: higher level information => piece together all available information - to get detailed picture of a molecular process (or disease) - to identif ...
A1991GD61600002
... on which it was based, summaDuring studies leading to the discovery rized these artides in a single location, and that vitamin Ba was a complex and that its included a summary of the metabolic inactive forms for lactic acid bacteria were terconversions leading from various forms pyridoxal and pyrido ...
... on which it was based, summaDuring studies leading to the discovery rized these artides in a single location, and that vitamin Ba was a complex and that its included a summary of the metabolic inactive forms for lactic acid bacteria were terconversions leading from various forms pyridoxal and pyrido ...
Enzymes - NVHSIntroBioPiper1
... Temperature (higher temperature = more activity until the enzyme’s protein denatures) ...
... Temperature (higher temperature = more activity until the enzyme’s protein denatures) ...
Bioinformatics - University of Oxford
... We can also identify the most likely mosaic structure and calculate the expected number of recombination events – Repeating the procedure for all sequences provides a way of assessing the importance of mosaicism within the family ...
... We can also identify the most likely mosaic structure and calculate the expected number of recombination events – Repeating the procedure for all sequences provides a way of assessing the importance of mosaicism within the family ...
Poster - Protein Information Resource
... confidently propagated from experimentallycharacterized proteins to uncharacterized proteins. The method relies upon rules that identify the specific amino acids in a protein chain eligible for tagging with appropriate information. Rules are specific for a particular protein family, and rely upon th ...
... confidently propagated from experimentallycharacterized proteins to uncharacterized proteins. The method relies upon rules that identify the specific amino acids in a protein chain eligible for tagging with appropriate information. Rules are specific for a particular protein family, and rely upon th ...
Chemical Reactions (L1)
... that are strong electrolytes (are soluble and dissociate) are written as their ions. – Some ions participate in the reaction – Some ions do NOT participate in the reaction-called spectator ...
... that are strong electrolytes (are soluble and dissociate) are written as their ions. – Some ions participate in the reaction – Some ions do NOT participate in the reaction-called spectator ...
Molecular Docking
... In the example below, the fit between the substrate and the protein is acceptable, except however for a bump with the phenyl group. The idea behind a soft scoring function is that instead of resolving bumps by conformational changes, it is possible to reduce their importance by using softer intera ...
... In the example below, the fit between the substrate and the protein is acceptable, except however for a bump with the phenyl group. The idea behind a soft scoring function is that instead of resolving bumps by conformational changes, it is possible to reduce their importance by using softer intera ...
template
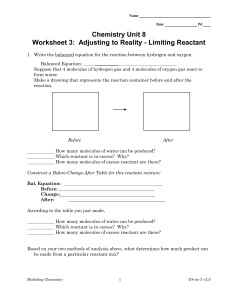
... 3. When 0.50 mole of aluminum reacts with 0.72 mole of iodine to form aluminum iodide, how many moles of the excess reactant will remain? ____________ How many moles of aluminum iodide will be formed? _____________ Bal. Equation: Before: __________________________________________ Change:___________ ...
... 3. When 0.50 mole of aluminum reacts with 0.72 mole of iodine to form aluminum iodide, how many moles of the excess reactant will remain? ____________ How many moles of aluminum iodide will be formed? _____________ Bal. Equation: Before: __________________________________________ Change:___________ ...
Modeling Membrane Movements
... Using the Membrane Model to Teach Transport Methods Simple Diffusion: 1. Put the model on the overhead projector. 2. Place the small marbles both inside the box ("cell") and outside the "cell" on the overhead transparency ("extracellular fluid"). Place many more on one side than the other. 3. Grab ...
... Using the Membrane Model to Teach Transport Methods Simple Diffusion: 1. Put the model on the overhead projector. 2. Place the small marbles both inside the box ("cell") and outside the "cell" on the overhead transparency ("extracellular fluid"). Place many more on one side than the other. 3. Grab ...
Types of Chemical Reactions (rxns.)
... Single Replacement Reactions: A metal can replace a metal (+) OR a nonmetal can replace a nonmetal (-). element + compoundà product + product A + BC à AC + B (if A is a metal) OR A + BC à BA + C (if A is a nonmetal) (remember the cation always goes first!) ...
... Single Replacement Reactions: A metal can replace a metal (+) OR a nonmetal can replace a nonmetal (-). element + compoundà product + product A + BC à AC + B (if A is a metal) OR A + BC à BA + C (if A is a nonmetal) (remember the cation always goes first!) ...
Project Advance Chemistry 106 Sample Questions
... respectively. Determine the value of the Eocell for a voltaic cell in which the overall reaction is ...
... respectively. Determine the value of the Eocell for a voltaic cell in which the overall reaction is ...
Nuclear Chemistry
... The fourth step is the six-carbon sugar fructose-1,6-diphosphate is split into two threecarbon compounds, dihydroxyacetone phosphate and glyceraldehyde-3-phosphae, catalyzed by the enzyme aldolase. ...
... The fourth step is the six-carbon sugar fructose-1,6-diphosphate is split into two threecarbon compounds, dihydroxyacetone phosphate and glyceraldehyde-3-phosphae, catalyzed by the enzyme aldolase. ...
INTRODUCTORY BIOCHEMISTRY Bio. 28 First Midterm
... 13. [4] Fill in the missing words: In the protein, __________________, additional strength is provided by the post-translational ____________________ of some amino acids to increase the number of __________________ bonds they can form. This modification becomes deficient when ____________________ is ...
... 13. [4] Fill in the missing words: In the protein, __________________, additional strength is provided by the post-translational ____________________ of some amino acids to increase the number of __________________ bonds they can form. This modification becomes deficient when ____________________ is ...
MolecularModelingDru..
... Outline Drug Discovery Approach • use the information in the databases and infer information that is not provided directly by genomics and proteomics data: higher level information => piece together all available information - to get detailed picture of a molecular process (or disease) - to identif ...
... Outline Drug Discovery Approach • use the information in the databases and infer information that is not provided directly by genomics and proteomics data: higher level information => piece together all available information - to get detailed picture of a molecular process (or disease) - to identif ...
View - University of Southampton
... electronic polarization contribution. With the addition of analytical first derivatives of eq 2 with respect to the nuclear coordinates, geometry optimizations can also be performed in the cavity, thus allowing for nuclear polarization effects as well. Two contributions that have been omitted from t ...
... electronic polarization contribution. With the addition of analytical first derivatives of eq 2 with respect to the nuclear coordinates, geometry optimizations can also be performed in the cavity, thus allowing for nuclear polarization effects as well. Two contributions that have been omitted from t ...
Dr. Audrey Lugo`s AP Chemistry Course Syllabus
... vaporization and fusion; calorimetry 3. Second law: entropy; free energy of formation; free energy of reaction; dependence of change in free energy on enthalpy and entropy changes 4. Relationship of change in free energy to equilibrium constants and electrode potentials IV. Descriptive Chemistry (10 ...
... vaporization and fusion; calorimetry 3. Second law: entropy; free energy of formation; free energy of reaction; dependence of change in free energy on enthalpy and entropy changes 4. Relationship of change in free energy to equilibrium constants and electrode potentials IV. Descriptive Chemistry (10 ...
igcse_enzyme_ppt
... Cheese is made by adding an enzyme called rennet after bacteria have produced lactic acid ...
... Cheese is made by adding an enzyme called rennet after bacteria have produced lactic acid ...
Chemical Equations and Reactions
... How to Balance an Equation 1. Balance the different types of atoms one at a time 2. First balance the atoms of elements that are combined and that appear only once on each side of the equation. (start with the largest compound first) 3. Balance polyatomic ions that appear on both sides of the equat ...
... How to Balance an Equation 1. Balance the different types of atoms one at a time 2. First balance the atoms of elements that are combined and that appear only once on each side of the equation. (start with the largest compound first) 3. Balance polyatomic ions that appear on both sides of the equat ...
ENZYME KINETICS - University of Pennsylvania
... disappearance of substrate or the appearance of product, depending upon which is chemically the more advantageous. The actual method of measurement depends on some biochemical or biophysical property of the molecule being assayed. For example, titration of acid or base or changes in absorption or ro ...
... disappearance of substrate or the appearance of product, depending upon which is chemically the more advantageous. The actual method of measurement depends on some biochemical or biophysical property of the molecule being assayed. For example, titration of acid or base or changes in absorption or ro ...
Simulated Pore Interactive Computing Environment (SPICE
... freedom which plays an important role in the IMD stage of the analysis and its use has been integrated into the RealityGrid framework. The device is connected to a controlling computer via an IEEE-1394 (’firewire’) link. The VMD10 visualisation package requires an additional software package, VRPN11 ...
... freedom which plays an important role in the IMD stage of the analysis and its use has been integrated into the RealityGrid framework. The device is connected to a controlling computer via an IEEE-1394 (’firewire’) link. The VMD10 visualisation package requires an additional software package, VRPN11 ...
Summer Work
... Fifth Exercise: The equilibrium constant for the reaction: 2 NO (g) N2 (g) + O2 (g) is 2.60 x 10-3 at 1100 °C. If 0.820 mole of NO (g) and 0.223 mole each of N2 (g) and O2 (g) are mixed in a 1.00 liter container at 1100 °C, what are the concentrations of NO (g), N2(g), and O2 (g) at equilibrium? ...
... Fifth Exercise: The equilibrium constant for the reaction: 2 NO (g) N2 (g) + O2 (g) is 2.60 x 10-3 at 1100 °C. If 0.820 mole of NO (g) and 0.223 mole each of N2 (g) and O2 (g) are mixed in a 1.00 liter container at 1100 °C, what are the concentrations of NO (g), N2(g), and O2 (g) at equilibrium? ...
Quiz - Columbus Labs
... Allosteric Regulation • allosteric means other site and an 'allosteric enzyme' is one with two binding sites - one for the substrate and one for the allosteric modifier molecule, which is not changed by the enzyme so it is not a substrate. • The molecule binding at the allosteric site is not called ...
... Allosteric Regulation • allosteric means other site and an 'allosteric enzyme' is one with two binding sites - one for the substrate and one for the allosteric modifier molecule, which is not changed by the enzyme so it is not a substrate. • The molecule binding at the allosteric site is not called ...
158KB - NZQA
... that the reaction lies to the products side as the larger the Kc or Q value, the greater the numerator (products). ...
... that the reaction lies to the products side as the larger the Kc or Q value, the greater the numerator (products). ...
Chemistry Syllabus
... 1st Nine Weeks 2a. Describe and classify matter based on physical and chemical properties and interactions between molecules or atoms. (DOK 1) Physical properties (e.g., melting points, densities, boiling points) of a variety of substances Substances and mixtures Three states of matter in term ...
... 1st Nine Weeks 2a. Describe and classify matter based on physical and chemical properties and interactions between molecules or atoms. (DOK 1) Physical properties (e.g., melting points, densities, boiling points) of a variety of substances Substances and mixtures Three states of matter in term ...