Definition (956.3 KB)
... The term protein quality refers to the ratio of essential amino acids (eaa) in a protein in comparison with the ratio required by the body. A high quality protein contains eaa in a ratio that matches human requirements. A protein which is lacking or low in one or more eaa is termed a low quality pro ...
... The term protein quality refers to the ratio of essential amino acids (eaa) in a protein in comparison with the ratio required by the body. A high quality protein contains eaa in a ratio that matches human requirements. A protein which is lacking or low in one or more eaa is termed a low quality pro ...
The Protein Interaction Prediction Engine (PIPE)
... Global Scan of Entire Protein Interaction Network species ...
... Global Scan of Entire Protein Interaction Network species ...
Supplementary method
... and 500mM NaCl). While conducting the assay, a no-peptide control is also included to account for any ATPase activity or auto-phosphorylation exhibited by the protein. The amount of protein used was optimized such that the kinase reaction was at least 25 times slower than the coupled PK/LDH reaction ...
... and 500mM NaCl). While conducting the assay, a no-peptide control is also included to account for any ATPase activity or auto-phosphorylation exhibited by the protein. The amount of protein used was optimized such that the kinase reaction was at least 25 times slower than the coupled PK/LDH reaction ...
ExoS binds its co-factor 14-3-3 through a non
... p186 replication by approximately 50-80%, while anti-14-3-3P antibodies had a lesser effect. All of these antibodies interfered with CBP’s binding t o cruciform D N A . The results indicate that CBP/14-3-3 is a replication origin binding protein, acting at the initiation step of D N A replication by ...
... p186 replication by approximately 50-80%, while anti-14-3-3P antibodies had a lesser effect. All of these antibodies interfered with CBP’s binding t o cruciform D N A . The results indicate that CBP/14-3-3 is a replication origin binding protein, acting at the initiation step of D N A replication by ...
Nitrogen Balance
... or structural protein • Essential amino acids in fish are the same as those in mammals: arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine (Halver and Shanks, 1960) • If the diet is deficient in lipid, then a greater proportion of dietary p ...
... or structural protein • Essential amino acids in fish are the same as those in mammals: arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine (Halver and Shanks, 1960) • If the diet is deficient in lipid, then a greater proportion of dietary p ...
Isotopica: a tool for the calculation and viewing of
... Received February 20, 2004; Revised and Accepted April 8, 2004 ...
... Received February 20, 2004; Revised and Accepted April 8, 2004 ...
Prof. Kamakaka`s Lecture 3 Notes
... (N–Ca—C–N) in the range ( 90° < y < 180°) • The planarity of the peptide bond and tetrahedral geometry of the a-carbon create a pleated sheetlike structure • Sheet-like arrangement of backbone is held together by hydrogen bonds between the more distal backbone amides • Side chains protrude from the ...
... (N–Ca—C–N) in the range ( 90° < y < 180°) • The planarity of the peptide bond and tetrahedral geometry of the a-carbon create a pleated sheetlike structure • Sheet-like arrangement of backbone is held together by hydrogen bonds between the more distal backbone amides • Side chains protrude from the ...
Grant Burgess
... NUCB (12-kda) eluted at the same position as cytochrome C (12.4kda). NUCB is monomeric. ...
... NUCB (12-kda) eluted at the same position as cytochrome C (12.4kda). NUCB is monomeric. ...
Poster Link
... amino acids in existence and proteins often consist of 300 or more amino acids. A “multiple alignment” is performed on a collection of sequences to maximize the areas where the amino acids are similar across all sequences. Online websites presently are available to accomplish the task. Once the mult ...
... amino acids in existence and proteins often consist of 300 or more amino acids. A “multiple alignment” is performed on a collection of sequences to maximize the areas where the amino acids are similar across all sequences. Online websites presently are available to accomplish the task. Once the mult ...
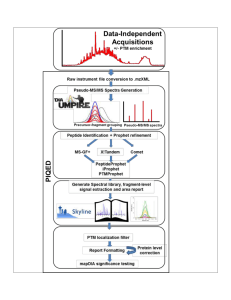
Supplementary Figure 1 Details of PIQED Automated Qualitative
... output from the previous module. These modules are broadly divided into three sections: (1) file conversions and DIAUmpire signal extraction, (2) database searches, and (3) post processing, quantification and significance testing. The leftmost section is used to specify parameters for file conversio ...
... output from the previous module. These modules are broadly divided into three sections: (1) file conversions and DIAUmpire signal extraction, (2) database searches, and (3) post processing, quantification and significance testing. The leftmost section is used to specify parameters for file conversio ...
Detection of Cellular Response to an in vitro Challenge with
... processed by Proteome Discoverer software using The Mascot® search engine. Two different peptide identification strategies were used. The simple search method (Figure 3) only searches for high-confidence, tryptic peptides and phosphopeptides. The more complex search strategy (Figure 4), breaks the P ...
... processed by Proteome Discoverer software using The Mascot® search engine. Two different peptide identification strategies were used. The simple search method (Figure 3) only searches for high-confidence, tryptic peptides and phosphopeptides. The more complex search strategy (Figure 4), breaks the P ...
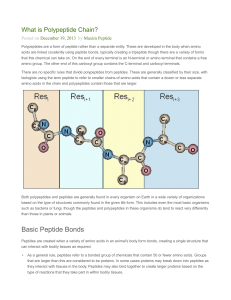
Basic Peptide Bonds
... that can be used to identify the protein structures involved in this process are still being investigated to better understand how this process works in the body naturally. The sequences of amino acids in polypeptides are controlled by codons which are located in the mRNA molecules that were used by ...
... that can be used to identify the protein structures involved in this process are still being investigated to better understand how this process works in the body naturally. The sequences of amino acids in polypeptides are controlled by codons which are located in the mRNA molecules that were used by ...
CSE182 project
... 5. Run BLASTX of the genomic queries against the non-redundant (nr) protein database. 6. For each translated region, determine if there exists a close protein homolog. If a homolog exists, then determine if peptides from the predicted region are supported by the alignment. Specifically, (a) Use the ...
... 5. Run BLASTX of the genomic queries against the non-redundant (nr) protein database. 6. For each translated region, determine if there exists a close protein homolog. If a homolog exists, then determine if peptides from the predicted region are supported by the alignment. Specifically, (a) Use the ...
Maintaining Linkage: More examples
... That is, sentences tend to sound like collections of stand-alone bullet points, because the connections between the ideas expressed in each sentence are not made clear. The result is a disjointed paragraph that readers have to work hard to follow. And if readers have to work too hard, they stop read ...
... That is, sentences tend to sound like collections of stand-alone bullet points, because the connections between the ideas expressed in each sentence are not made clear. The result is a disjointed paragraph that readers have to work hard to follow. And if readers have to work too hard, they stop read ...
Amino acids and Protein Structure
... Protein subunits: amino acids: L & D isomers Only L amino acids found in proteins. C chiral, L & D isomers not symmetrical, except glycine R group = side chains ...
... Protein subunits: amino acids: L & D isomers Only L amino acids found in proteins. C chiral, L & D isomers not symmetrical, except glycine R group = side chains ...
MALDI-MS
... Biomolecules and organic macromolecules are fragile Molecular ions or meaningful fragments were limited to only 5- ...
... Biomolecules and organic macromolecules are fragile Molecular ions or meaningful fragments were limited to only 5- ...
The Subcellular Distribution of Multigene Family 110 Proteins of
... region of the genome expressing MGF genes (14), suggesting that maintenance of MGFs may be important for the propagation of ASFV in the tick vector. The cellular functions of most MGF proteins are largely unknown and difficult to predict because the proteins show limited homology to host proteins. F ...
... region of the genome expressing MGF genes (14), suggesting that maintenance of MGFs may be important for the propagation of ASFV in the tick vector. The cellular functions of most MGF proteins are largely unknown and difficult to predict because the proteins show limited homology to host proteins. F ...
生物物理学 I Handout No. 2 ① ② ③ ④ ⑤
... between the two states is proposed to occur randomly and to be completely reversible. Therefore, if the concentration of A is higher on the outside of the bilayer, more A will bind to the carrier protein in the pong conformation than in the ping conformation, and there will be a net transport of A d ...
... between the two states is proposed to occur randomly and to be completely reversible. Therefore, if the concentration of A is higher on the outside of the bilayer, more A will bind to the carrier protein in the pong conformation than in the ping conformation, and there will be a net transport of A d ...
: BIOGRAPHICAL SKETCH
... potential diagnostic and prognostic biomarkers to develop clinical diagnostics as a part of the system biology approach. ...
... potential diagnostic and prognostic biomarkers to develop clinical diagnostics as a part of the system biology approach. ...
AP Biology Test 1 Organic Chemistry Part III. Organic Molecules 1
... protein with tertiary structure. B) lipid made with three fatty acids and glycerol. C) lipid that makes up much of the plasma membrane. D) molecule formed from three alcohols by dehydration reactions. E) carbohydrate with three sugars joined together by glycosidic linkages. ...
... protein with tertiary structure. B) lipid made with three fatty acids and glycerol. C) lipid that makes up much of the plasma membrane. D) molecule formed from three alcohols by dehydration reactions. E) carbohydrate with three sugars joined together by glycosidic linkages. ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.