Over Expression of IPTG inducible GST protein in E.coli BL21
... these enzyme involves in nucleophilic attack by glutathione on an electrophilic substrate [2]. The resulting glutathione S conjugate are more soluble than the original substrate and thus more easily transported from the cell, mediated by ATP dependent MAPEG family membrane glycoprotein belonging to ...
... these enzyme involves in nucleophilic attack by glutathione on an electrophilic substrate [2]. The resulting glutathione S conjugate are more soluble than the original substrate and thus more easily transported from the cell, mediated by ATP dependent MAPEG family membrane glycoprotein belonging to ...
Protein Creation Pathway Tutorial
... Directions: Download and then open the PowerPoint from my website. Press the F5 button to start the presentation. ...
... Directions: Download and then open the PowerPoint from my website. Press the F5 button to start the presentation. ...
Supplementary information
... being washed three times with TBST (0.02% Tween 20 in TBS) for 10 min, the membrane was incubated in alkaline phosphatase-conjugated goat-anti-rabbit IgG (1:10,000 in the blocking solution) for 3 h. The membrane was washed three times with TBST for 10 min each and once in TBS for 10 min. The signal ...
... being washed three times with TBST (0.02% Tween 20 in TBS) for 10 min, the membrane was incubated in alkaline phosphatase-conjugated goat-anti-rabbit IgG (1:10,000 in the blocking solution) for 3 h. The membrane was washed three times with TBST for 10 min each and once in TBS for 10 min. The signal ...
sample - Create Training
... Nutrients are divided into major and minor nutrients. Major nutrients, which are carbohydrates, proteins, and lipids (fats), are used as energy sources or as building blocks for larger biochemical compounds. Minor nutrients, which include all vitamins and minerals, assist the chemical reactions that ...
... Nutrients are divided into major and minor nutrients. Major nutrients, which are carbohydrates, proteins, and lipids (fats), are used as energy sources or as building blocks for larger biochemical compounds. Minor nutrients, which include all vitamins and minerals, assist the chemical reactions that ...
Poster for RCPSC mee.. - University of Alberta
... (GlcCer) is a metabolite of ceramide produced by the glycosylation of the 1-hydroxyl group of ceramide by the enzyme Glucosylceramide Synthase (GCS) (Figure 1). Given the similarities in structure between the natural product inhibitors of PP1, the clavosines, and the sphingolipid GlcCer, we hypothes ...
... (GlcCer) is a metabolite of ceramide produced by the glycosylation of the 1-hydroxyl group of ceramide by the enzyme Glucosylceramide Synthase (GCS) (Figure 1). Given the similarities in structure between the natural product inhibitors of PP1, the clavosines, and the sphingolipid GlcCer, we hypothes ...
A Lipid Droplet Protein of Nannochloropsis with
... of N deprivation (Cell N-), and the LD fraction (LD). Arrow indicates 17-kD protein submitted to mass spectrometry. C, Kyte-Doolittle hydropathy index (HI) with window size of 9; r.U., relative units. ...
... of N deprivation (Cell N-), and the LD fraction (LD). Arrow indicates 17-kD protein submitted to mass spectrometry. C, Kyte-Doolittle hydropathy index (HI) with window size of 9; r.U., relative units. ...
Journal of Agricultural and Food Chemistry
... Caryophyllales (e.g., quinoa and amaranth) are closely related. Our data rather indicate occurrence of significant genetic distance (similarity coefficients 0.05-0.10). Also, it is doubtful that amaranth and quinoa species are also closely related (similarity coefficients varied from 0.16 to 0.25). ...
... Caryophyllales (e.g., quinoa and amaranth) are closely related. Our data rather indicate occurrence of significant genetic distance (similarity coefficients 0.05-0.10). Also, it is doubtful that amaranth and quinoa species are also closely related (similarity coefficients varied from 0.16 to 0.25). ...
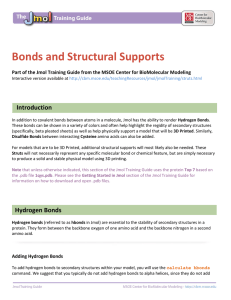
Bonds and Structural Supports - MSOE Center for BioMolecular
... When a model is built on a 3D printer, additional support struts are added within the model to provide stability and durability. Struts can be automatically added using the calculate struts command. This command will only add struts to the currently selected area of a molecular structure. If you hav ...
... When a model is built on a 3D printer, additional support struts are added within the model to provide stability and durability. Struts can be automatically added using the calculate struts command. This command will only add struts to the currently selected area of a molecular structure. If you hav ...
CARBOHYDRATES 2016
... CONFIGURATION D and L Molecules of glyceric aldehyde are enantiomers. Stereoisomers are isomeric molecules that have the same molecular formula and sequence of bonded atoms (constitution), but which differ only in the three-dimensional orientations of their atoms in space. Also are called mirror im ...
... CONFIGURATION D and L Molecules of glyceric aldehyde are enantiomers. Stereoisomers are isomeric molecules that have the same molecular formula and sequence of bonded atoms (constitution), but which differ only in the three-dimensional orientations of their atoms in space. Also are called mirror im ...
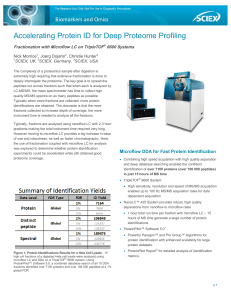
Accelerating Protein ID for Deep Proteome Profiling
... higher quality SWATH® acquisition library. ...
... higher quality SWATH® acquisition library. ...
Database searching with DNA and protein sequences
... on observed substitution schemes – tends to give more accurate matches for distantly related sequences. These matrices are derived by analysing how often one amino acid is seen to substitute for another in alignments of well-characterised protein families. One feature of this type of scheme is that ...
... on observed substitution schemes – tends to give more accurate matches for distantly related sequences. These matrices are derived by analysing how often one amino acid is seen to substitute for another in alignments of well-characterised protein families. One feature of this type of scheme is that ...
8.5 Translation - Cloudfront.net
... • Translation converts mRNA messages into polypeptides which make ...
... • Translation converts mRNA messages into polypeptides which make ...
View PDF - Sutro Biopharma, Inc.
... More recently, the PURE system has enabled reprogramming of the genetic code with simultaneous complete reassignment of multiple codons to different nnAAs. By removing certain cognate amino acids and AARSs and adding separately nnAA aminoacylated-tRNAs to recognize the vacant codons, the translation ...
... More recently, the PURE system has enabled reprogramming of the genetic code with simultaneous complete reassignment of multiple codons to different nnAAs. By removing certain cognate amino acids and AARSs and adding separately nnAA aminoacylated-tRNAs to recognize the vacant codons, the translation ...
02_Murray - Sbkb.org
... Collaborations with Experimental Groups Characterize different START domains based on structural information Discriminate whether START domains bind cholesterol or PC (PI) or other ligands Provide leads for chemical library studies for function-interfering compounds Detailed computational analysis ...
... Collaborations with Experimental Groups Characterize different START domains based on structural information Discriminate whether START domains bind cholesterol or PC (PI) or other ligands Provide leads for chemical library studies for function-interfering compounds Detailed computational analysis ...
BMS 6204 MEDICAL BIOCHEMISTRY & GENETICS SPRING 2010
... 3.6 ATP: Do I really want that second serving of pasta? Let's do the math (anaerobic vs. aerobic) 3.7 Glucose Storage: glycogenesis/glycogenolysis Small groups (3.1) 3.8 Gluconeogenesis: Glucogenic and ketogenic amino acids Muscle Metabolism: Cori cycle 3.9 Urea Synthesis and Metabolism Q & A with L ...
... 3.6 ATP: Do I really want that second serving of pasta? Let's do the math (anaerobic vs. aerobic) 3.7 Glucose Storage: glycogenesis/glycogenolysis Small groups (3.1) 3.8 Gluconeogenesis: Glucogenic and ketogenic amino acids Muscle Metabolism: Cori cycle 3.9 Urea Synthesis and Metabolism Q & A with L ...
Relative Requirements for Magnesium of Protein and Chlorophyll
... Kinetics of Growth and Formation of Chi and protein. When photoheterotrophic Euglena are grown with different amounts of Mg initially present in the medium, both the rate of growth and total yield, as measured by apparent optical density, are affected (Fig. IA). Provided that exponentially growing c ...
... Kinetics of Growth and Formation of Chi and protein. When photoheterotrophic Euglena are grown with different amounts of Mg initially present in the medium, both the rate of growth and total yield, as measured by apparent optical density, are affected (Fig. IA). Provided that exponentially growing c ...
Laboratory Exercise #7: Column Chromatography of GFP proteins
... acids. In salt water, these parts of the proteins will adhere to other hydrophobic substances. So a high salt concentration will cause the 3D structure of a protein to change so that the hydrophobic regions of the protein are exposed on the surface of the protein and the water-loving hydrophilic re ...
... acids. In salt water, these parts of the proteins will adhere to other hydrophobic substances. So a high salt concentration will cause the 3D structure of a protein to change so that the hydrophobic regions of the protein are exposed on the surface of the protein and the water-loving hydrophilic re ...
Word document
... enzymes for Agrobacterium tumefaciens. The other key enzyme of this pathway which was detected highly abundant in the pH 7 mass spectrometry data set is Transketolase. You become curious about the nature of this protein and start to explore it a bit more, e.g. in order to have enough structure-funct ...
... enzymes for Agrobacterium tumefaciens. The other key enzyme of this pathway which was detected highly abundant in the pH 7 mass spectrometry data set is Transketolase. You become curious about the nature of this protein and start to explore it a bit more, e.g. in order to have enough structure-funct ...
supp - Springer Static Content Server
... HnRNP F is an RNA binding protein belonging to the HnRNP H protein family that contains four highly homologous members, namely HnRNP H, HnRNP H’, HnRNP F, and HnRNP 2H9. These proteins contain two (2H9) or three (H, H’, and F) RNA recognition motifs (RRM) and two glycine rich auxiliary domains (Hono ...
... HnRNP F is an RNA binding protein belonging to the HnRNP H protein family that contains four highly homologous members, namely HnRNP H, HnRNP H’, HnRNP F, and HnRNP 2H9. These proteins contain two (2H9) or three (H, H’, and F) RNA recognition motifs (RRM) and two glycine rich auxiliary domains (Hono ...
Increasing Muscle Growth
... individual may need based on their resistance digesting, leucine-rich protein, such as whey. training is not an easy task. Each individual is This same study also looked into the differences different and reacts differently to both training and between whey, soy, and casein on the average ingesting ...
... individual may need based on their resistance digesting, leucine-rich protein, such as whey. training is not an easy task. Each individual is This same study also looked into the differences different and reacts differently to both training and between whey, soy, and casein on the average ingesting ...
Title: Rescuing discarded spectra: Full - e
... 040 were assigned to peptide sequences (73%; Figures 1B and S1) with a FDR of 5%. However, only 35% of these (17 817) were matched to sequences present in the NCBInr database (Supporting Information Table S1; Figures 1B and S1), and of those, 14 239 PSMs were associated to 496 M. pneumoniae proteins ...
... 040 were assigned to peptide sequences (73%; Figures 1B and S1) with a FDR of 5%. However, only 35% of these (17 817) were matched to sequences present in the NCBInr database (Supporting Information Table S1; Figures 1B and S1), and of those, 14 239 PSMs were associated to 496 M. pneumoniae proteins ...
Protein structure prediction

Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its folding and its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.