Chapter 21 - HCC Learning Web
... • Using available DNA sequences, geneticists can study genes directly in an approach called reverse genetics • The identification of protein coding genes within DNA sequences in a database is called gene annotation • Gene annotation is largely an automated process • Comparison of sequences of previo ...
... • Using available DNA sequences, geneticists can study genes directly in an approach called reverse genetics • The identification of protein coding genes within DNA sequences in a database is called gene annotation • Gene annotation is largely an automated process • Comparison of sequences of previo ...
Lecture 3 UG
... Different cells have different sets of receptor for the same ligand and each of which induces a different response Different cells respond in a variety of way to the same ligand (e.g. acetylcholine) Different ligands can induce the same cellular response in some cells (glucagon/epinephrine) In most ...
... Different cells have different sets of receptor for the same ligand and each of which induces a different response Different cells respond in a variety of way to the same ligand (e.g. acetylcholine) Different ligands can induce the same cellular response in some cells (glucagon/epinephrine) In most ...
Study Guide MBMB 451A Fall 2002
... 7. What is an enhancer? What is a response element? 8. Describe two models for how an enhancer could effect the level of transcription. 9. What are the transcription factors called that are used by Pol I and Pol III? 10. Discuss how transcription activity can be regulated by protein phosphorylation ...
... 7. What is an enhancer? What is a response element? 8. Describe two models for how an enhancer could effect the level of transcription. 9. What are the transcription factors called that are used by Pol I and Pol III? 10. Discuss how transcription activity can be regulated by protein phosphorylation ...
Bio200 Au13 Lec19 10-29 Slides
... • Eukaryotic genes can have promoter-influencing elements that are far upstream or downstream of the protein-coding region. • These silencers and enhancers work through DNA binding proteins that either help to recruit to repel RNA polymerase. DNA flexibility is essential. ...
... • Eukaryotic genes can have promoter-influencing elements that are far upstream or downstream of the protein-coding region. • These silencers and enhancers work through DNA binding proteins that either help to recruit to repel RNA polymerase. DNA flexibility is essential. ...
PDF
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
Unit 3 Concepts Study Guide
... 5. Scientists calculate the similarities of gene expression patterns between different individuals using statistical analysis. - Correlational coefficients: if the value is close to zero (little to no correlation); close to +1 (gene expression is similar); close to -1 (gene expression is opposite). ...
... 5. Scientists calculate the similarities of gene expression patterns between different individuals using statistical analysis. - Correlational coefficients: if the value is close to zero (little to no correlation); close to +1 (gene expression is similar); close to -1 (gene expression is opposite). ...
PDF
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
PDF
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
... requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (p. 1863). Their result ...
Regulation of gene expression
... Genetic regulation • Genotype is not phenotype: bacteria possess many genes that they are not using at any particular time. • Transcription and translation are expensive; why spend ATP to make an enzyme you don’t need? • Operon – Genes physically adjacent regulated together ...
... Genetic regulation • Genotype is not phenotype: bacteria possess many genes that they are not using at any particular time. • Transcription and translation are expensive; why spend ATP to make an enzyme you don’t need? • Operon – Genes physically adjacent regulated together ...
DNA methylation signature of activated human natural killer cells
... cluster important in Th2 activation. The six probes from IL5 and the nine probes from IL13 were manually clustered by NK activation status and assessed for methylation status. A t-test was performed for each gene to compare naïve (T = 0) NKs with activated (T = 9) NKs, and p-values computed. The ast ...
... cluster important in Th2 activation. The six probes from IL5 and the nine probes from IL13 were manually clustered by NK activation status and assessed for methylation status. A t-test was performed for each gene to compare naïve (T = 0) NKs with activated (T = 9) NKs, and p-values computed. The ast ...
One copy from each parent Each parent passes on a “mixed copy”
... Molecular Cell Biology: Components of the Central Dogma Protein Translation ...
... Molecular Cell Biology: Components of the Central Dogma Protein Translation ...
Biotechnology Notes
... – can detect genes related to an increased risk of cancer – can detect some genes known to cause genetic disorders ...
... – can detect genes related to an increased risk of cancer – can detect some genes known to cause genetic disorders ...
View PDF - Genetics
... The developmental complexity of multicellular organisms can obscure whether phenotypes resulting from loss of a ubiquitous regulator reflect the role of that factor in every tissue or only one. Carter et al. demonstrate that multiple reproductive defects associated with loss of a CHD chromatin remod ...
... The developmental complexity of multicellular organisms can obscure whether phenotypes resulting from loss of a ubiquitous regulator reflect the role of that factor in every tissue or only one. Carter et al. demonstrate that multiple reproductive defects associated with loss of a CHD chromatin remod ...
bioinformatix-ex
... dot on a chip measures the expression of one gene under a given environment condition. The output data is stored for each gene with experiment context and reference information usually from public sources. Commonly used analysis methods include statistical analysis (hypothesis testing), visualisatio ...
... dot on a chip measures the expression of one gene under a given environment condition. The output data is stored for each gene with experiment context and reference information usually from public sources. Commonly used analysis methods include statistical analysis (hypothesis testing), visualisatio ...
Homer-Wright rosettes
... Incidence of malignancy:1-15 yrs 1.3 /10,000 /year but leading cause of death after accidents in the West. (developing countries??) Most malignant tumours in children arise from hematopoietic,nervous and soft ...
... Incidence of malignancy:1-15 yrs 1.3 /10,000 /year but leading cause of death after accidents in the West. (developing countries??) Most malignant tumours in children arise from hematopoietic,nervous and soft ...
Biology 105: Biology Science for Life with Physiology, 3rd Ed., Belk
... 8 complementary base pair; 9 codon;10 degenerative diseases; 11 deoxyribose; 12 DNA polymerase; 13 DNA replication; 14 frameshift mutation;15 galls;16 germ-line gene therapy; 17 gene gun; 18 gene therapy; 19 generally recognized as safe (GRAS); 20 genetically modified organism (GMO); 21 genetic code ...
... 8 complementary base pair; 9 codon;10 degenerative diseases; 11 deoxyribose; 12 DNA polymerase; 13 DNA replication; 14 frameshift mutation;15 galls;16 germ-line gene therapy; 17 gene gun; 18 gene therapy; 19 generally recognized as safe (GRAS); 20 genetically modified organism (GMO); 21 genetic code ...
Development and Apoptosis
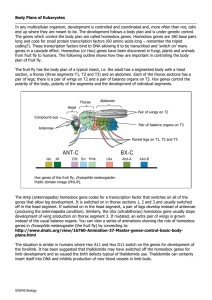
... Body Plans of Eukaryotes In any multicellular organism, development is controlled and coordinated and, more often than not, cells end up where they are meant to be. The development follows a body plan and is under genetic control. The genes which control the body plan are called homeobox genes. Home ...
... Body Plans of Eukaryotes In any multicellular organism, development is controlled and coordinated and, more often than not, cells end up where they are meant to be. The development follows a body plan and is under genetic control. The genes which control the body plan are called homeobox genes. Home ...
MBLG2x71 Course Information for mmb web site
... 14. Review. Review of the themes of the last 6 lectures, covering exam style questions. 15. Introduction to the structure of the Genome Review DNA structure with A, B and Z of DNA. DNA packging. Chromosome length and diversity, differences between eukaryotic and prokaryotic chromosomes, packaging pr ...
... 14. Review. Review of the themes of the last 6 lectures, covering exam style questions. 15. Introduction to the structure of the Genome Review DNA structure with A, B and Z of DNA. DNA packging. Chromosome length and diversity, differences between eukaryotic and prokaryotic chromosomes, packaging pr ...
Identification of Mucin 2 as a Strong Promoter for Gut
... of gut-specific genes could significantly improve poultry production. In the present study, the mucin 2 promoter is used to drive overexpression of green florescent protein (GFP) in intestinal tissue. Through comparison of gene expression in different tissues, the gut specificity of mucin 2 expressi ...
... of gut-specific genes could significantly improve poultry production. In the present study, the mucin 2 promoter is used to drive overexpression of green florescent protein (GFP) in intestinal tissue. Through comparison of gene expression in different tissues, the gut specificity of mucin 2 expressi ...
Ch. 16 – Control of Gene Expression Sample Questions
... A.Have their transcription occurring in the cytoplasm and translation in the nucleus. B.Have their transcription occurring in the nucleus and translation in the cytoplasm. C.Have only operons to assist in gene expression. D.Carry out protein synthesis only in the presence of the cAMP molecule. E.Use ...
... A.Have their transcription occurring in the cytoplasm and translation in the nucleus. B.Have their transcription occurring in the nucleus and translation in the cytoplasm. C.Have only operons to assist in gene expression. D.Carry out protein synthesis only in the presence of the cAMP molecule. E.Use ...
pGLO Transformation Lab Background Information Introduction to
... (catabolism) of food are good examples of highly regulated genes. For example, the sugar arabinose is both a source of energy and a source of carbon. E. coli bacteria produce three enzymes (proteins) needed to digest arabinose as a food source. The genes which code for these enzymes are not expresse ...
... (catabolism) of food are good examples of highly regulated genes. For example, the sugar arabinose is both a source of energy and a source of carbon. E. coli bacteria produce three enzymes (proteins) needed to digest arabinose as a food source. The genes which code for these enzymes are not expresse ...
2. The drug development process
... Biological function of between one-third and half of sequenced gene products remains unknown Assessment of biological functions of the sequenced genes Crucial to understanding the relationship between genotype and phenotype as well as direct identification of drug targets Shift in the focus ...
... Biological function of between one-third and half of sequenced gene products remains unknown Assessment of biological functions of the sequenced genes Crucial to understanding the relationship between genotype and phenotype as well as direct identification of drug targets Shift in the focus ...
genomebiology.com
... level) but also its kinetics: the gene associated with the CMV promoter remains on at all times, with a high number of engaged polymerases. The CCND1 promoter, in contrast, alternates between ON and OFF states, with an overall lower number of engaged polymerases. These bursts are not necessarily inc ...
... level) but also its kinetics: the gene associated with the CMV promoter remains on at all times, with a high number of engaged polymerases. The CCND1 promoter, in contrast, alternates between ON and OFF states, with an overall lower number of engaged polymerases. These bursts are not necessarily inc ...
RNA Synthetic Biology
... Rackham, Oliver, and Jason W. Chin. "A Network of Orthogonal RibosomemRNA Pairs." Nature Chemical Biotechnology 1.3 (2005): 159-66. Rackham, O. & Chin, J.W. Cellular logic with orthogonal ribosomes. Journal of the Americal Chemical Society 127, 17584–17585 (2005). Stojanovic, M.N. & Stefanovic, D. A ...
... Rackham, Oliver, and Jason W. Chin. "A Network of Orthogonal RibosomemRNA Pairs." Nature Chemical Biotechnology 1.3 (2005): 159-66. Rackham, O. & Chin, J.W. Cellular logic with orthogonal ribosomes. Journal of the Americal Chemical Society 127, 17584–17585 (2005). Stojanovic, M.N. & Stefanovic, D. A ...
Gene regulatory network

A gene regulatory network or genetic regulatory network (GRN) is a collection of regulators thatinteract with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins.The regulator can be DNA, RNA, protein and their complex. The interaction can be direct or indirect (through their transcribed RNA or translated protein).In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enzyme, i.e., a micro-machine that catalyses a certain reaction, such as the breakdown of a food source or toxin. Some proteins though serve only to activate other genes, and these are the transcription factors that are the main players in regulatory networks or cascades. By binding to the promoter region at the start of other genes they turn them on, initiating the production of another protein, and so on. Some transcription factors are inhibitory.In single-celled organisms, regulatory networks respond to the external environment, optimising the cell at a given time for survival in this environment. Thus a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol. This process, which we associate with wine-making, is how the yeast cell makes its living, gaining energy to multiply, which under normal circumstances would enhance its survival prospects.In multicellular animals the same principle has been put in the service of gene cascades that control body-shape. Each time a cell divides, two cells result which, although they contain the same genome in full, can differ in which genes are turned on and making proteins. Sometimes a 'self-sustaining feedback loop' ensures that a cell maintains its identity and passes it on. Less understood is the mechanism of epigenetics by which chromatin modification may provide cellular memory by blocking or allowing transcription. A major feature of multicellular animals is the use of morphogen gradients, which in effect provide a positioning system that tells a cell where in the body it is, and hence what sort of cell to become. A gene that is turned on in one cell may make a product that leaves the cell and diffuses through adjacent cells, entering them and turning on genes only when it is present above a certain threshold level. These cells are thus induced into a new fate, and may even generate other morphogens that signal back to the original cell. Over longer distances morphogens may use the active process of signal transduction. Such signalling controls embryogenesis, the building of a body plan from scratch through a series of sequential steps. They also control and maintain adult bodies through feedback processes, and the loss of such feedback because of a mutation can be responsible for the cell proliferation that is seen in cancer. In parallel with this process of building structure, the gene cascade turns on genes that make structural proteins that give each cell the physical properties it needs.It has been suggested that, because biological molecular interactions are intrinsically stochastic, gene networks are the result of cellular processes and not their cause (i.e. cellular Darwinism). However, recent experimental evidence has favored the attractor view of cell fates.