Previous IB Exam Essay Questions: Basic Molecules, Proteins
... annotated graph showing reaction with and without enzyme; substrate joins with enzyme at active site; to form enzyme-substrate complex; active site/enzyme (usually) specific for a particular substrate; enzyme binding with substrate brings reactants closer together to facilitate chemical reactions (s ...
... annotated graph showing reaction with and without enzyme; substrate joins with enzyme at active site; to form enzyme-substrate complex; active site/enzyme (usually) specific for a particular substrate; enzyme binding with substrate brings reactants closer together to facilitate chemical reactions (s ...
Slide 1
... (A) The structure of a fragment of a mouse gene regulatory protein bound to a specific DNA site. This protein recognizes DNA using three zinc fingers of the Cys-Cys-His-His type arranged as direct repeats. (B) The three fingers have similar amino acid sequences and contact the DNA in similar ways. I ...
... (A) The structure of a fragment of a mouse gene regulatory protein bound to a specific DNA site. This protein recognizes DNA using three zinc fingers of the Cys-Cys-His-His type arranged as direct repeats. (B) The three fingers have similar amino acid sequences and contact the DNA in similar ways. I ...
Crossing Membranes – Passive Processes
... molecule can bind causing a change in shape of the protein that delivers the molecule to the other side of the membrane. It may saturate when all the carrier proteins are in use ...
... molecule can bind causing a change in shape of the protein that delivers the molecule to the other side of the membrane. It may saturate when all the carrier proteins are in use ...
Protein Structure Prediction and Display
... Proteins with similar structures are termed “VAST Neighbors” by Entrez (VAST refers to the method used to evaluate similarity of structure) VAST or structure neighbors may or may not have sequence homology to each other ...
... Proteins with similar structures are termed “VAST Neighbors” by Entrez (VAST refers to the method used to evaluate similarity of structure) VAST or structure neighbors may or may not have sequence homology to each other ...
Protein folding: mechanisms and role in disease - Max
... the hippocampus, disturbing the complex neural networks of this brain region, resulting in cell death and loss of memory function (Figure 4A). It turns out that a particular form of protein aggregate is most intimately involved with these toxic phenomena. These aggregates are highly ordered fibrils ...
... the hippocampus, disturbing the complex neural networks of this brain region, resulting in cell death and loss of memory function (Figure 4A). It turns out that a particular form of protein aggregate is most intimately involved with these toxic phenomena. These aggregates are highly ordered fibrils ...
Expediting Purification of Cellular Proteins
... molecules or cellular events to global functional analysis, feeding these results into new approaches for the prevention, diagnosis, and treatment of cancer. Methods that allow researchers to look across a broader angle at cellular processes such as mRNA expression levels or protein interaction patt ...
... molecules or cellular events to global functional analysis, feeding these results into new approaches for the prevention, diagnosis, and treatment of cancer. Methods that allow researchers to look across a broader angle at cellular processes such as mRNA expression levels or protein interaction patt ...
Chapter 14
... Ribosomes have two subunits, large and small, held together noncovalently. In eukaryotes, the large subunit has three different molecules of ribosomal RNA (rRNA) and 49 different proteins in a precise pattern. The small subunit has one rRNA and ...
... Ribosomes have two subunits, large and small, held together noncovalently. In eukaryotes, the large subunit has three different molecules of ribosomal RNA (rRNA) and 49 different proteins in a precise pattern. The small subunit has one rRNA and ...
2016-10-12 Jurgen Chemical Proteomics
... … aims to study how small molecules (“chemicals”) of synthetic or natural origin bind to proteins and modulate their function. … can be applied in drug target discovery or to identify small-molecule probes as research tools to study protein function. … often relies on current state-of-the-art in pro ...
... … aims to study how small molecules (“chemicals”) of synthetic or natural origin bind to proteins and modulate their function. … can be applied in drug target discovery or to identify small-molecule probes as research tools to study protein function. … often relies on current state-of-the-art in pro ...
File
... There are several types of lipids, but all contain subunits of glycerol and fatty acids made of carbon, hydrogen, and oxygen. It is different from a carbohydrate because of the ratio and because the smaller units do not link together to form a chemical chain ...
... There are several types of lipids, but all contain subunits of glycerol and fatty acids made of carbon, hydrogen, and oxygen. It is different from a carbohydrate because of the ratio and because the smaller units do not link together to form a chemical chain ...
Prediction for Essential Proteins with the Support Vector Machine
... protein does not have a unique score. Thus, we propose a bit string implementation to transform the ranking to a score of protein. We select two topological properties for ranking, A and B. For the n target proteins, we rank them by A and B individually. Then, n2 iterations are performed to construc ...
... protein does not have a unique score. Thus, we propose a bit string implementation to transform the ranking to a score of protein. We select two topological properties for ranking, A and B. For the n target proteins, we rank them by A and B individually. Then, n2 iterations are performed to construc ...
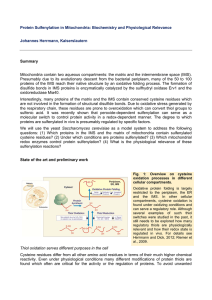
Protein Sulfenylation in Mitochondria: Biochemistry and
... so that at least under non-stressed conditions most protein thiols are reduced (Grant, 2001; Hansen et al., 2009; Kumar et al., 2011). Nevertheless, thiol oxidation can be used here in order to sense environmental conditions such as the concentrations of oxygen or hydrogen peroxide. For example, in ...
... so that at least under non-stressed conditions most protein thiols are reduced (Grant, 2001; Hansen et al., 2009; Kumar et al., 2011). Nevertheless, thiol oxidation can be used here in order to sense environmental conditions such as the concentrations of oxygen or hydrogen peroxide. For example, in ...
Rampant Purifying Selection Conserves Positions with
... 7 times greater than the slowest 25% evolving proteins. Overall, phosphorylated proteins are more conserved than N-linked glycosylated proteins (20% difference; P ,, 0.01; t-test with unequal variances). Similarly, different amino acid residues evolve with different rates (fig. 2), with tyrosines sh ...
... 7 times greater than the slowest 25% evolving proteins. Overall, phosphorylated proteins are more conserved than N-linked glycosylated proteins (20% difference; P ,, 0.01; t-test with unequal variances). Similarly, different amino acid residues evolve with different rates (fig. 2), with tyrosines sh ...
Investigating the Role of RNA Polymerase II in RNAi
... methyltransferase that catalyzes addition of the H3K9 methyl mark), which leads to histone modification and ultimately gene silencing (Bühler and Moazed, 2007). Much has been learned about this mechanism. Not surprisingly, deletion mutants for parts of the histone-modifying CLRC complex fail to exhi ...
... methyltransferase that catalyzes addition of the H3K9 methyl mark), which leads to histone modification and ultimately gene silencing (Bühler and Moazed, 2007). Much has been learned about this mechanism. Not surprisingly, deletion mutants for parts of the histone-modifying CLRC complex fail to exhi ...
B Ca(2+)
... at 12,000X magnification on a JEOL 2000 EX-II 200 keV transmission electron microscope. Images were scanned using an Agfa DuoScan T2500 and adjusted for contrast using Adobe Photoshop software. ...
... at 12,000X magnification on a JEOL 2000 EX-II 200 keV transmission electron microscope. Images were scanned using an Agfa DuoScan T2500 and adjusted for contrast using Adobe Photoshop software. ...
Untitled
... hydrophobic so that a water/organic solvent mobile phase is used, that is, the stationary phase is more hydrophobic than the mobile phase. RPC media may be referred to as adsorbents while eluent solutions may be referred to as mobile phases. The separation of biomolecules by RPC depends on a reversi ...
... hydrophobic so that a water/organic solvent mobile phase is used, that is, the stationary phase is more hydrophobic than the mobile phase. RPC media may be referred to as adsorbents while eluent solutions may be referred to as mobile phases. The separation of biomolecules by RPC depends on a reversi ...
8.4 Transcription
... • RNA Polymerase • Enzyme that catalyzes the synthesis of a complementary strand of RNA from a DNA template. • Enzymes that bond nucleotides together in a chain to make a new RNA molecule. • Messenger RNA (mRNA) • Form of RNA that carries genetic information from the nucleus to the cytoplasm, where ...
... • RNA Polymerase • Enzyme that catalyzes the synthesis of a complementary strand of RNA from a DNA template. • Enzymes that bond nucleotides together in a chain to make a new RNA molecule. • Messenger RNA (mRNA) • Form of RNA that carries genetic information from the nucleus to the cytoplasm, where ...
Chapt21 Lecture 13ed Pt 2
... ________ while RNA is ______________. – DNA has T while RNA has U. – RNA is also found in the ___________ as well as the nucleus while DNA is not. ...
... ________ while RNA is ______________. – DNA has T while RNA has U. – RNA is also found in the ___________ as well as the nucleus while DNA is not. ...
8.4 Transcription
... • RNA Polymerase • Enzyme that catalyzes the synthesis of a complementary strand of RNA from a DNA template. • Enzymes that bond nucleotides together in a chain to make a new RNA molecule. • Messenger RNA (mRNA) • Form of RNA that carries genetic information from the nucleus to the cytoplasm, where ...
... • RNA Polymerase • Enzyme that catalyzes the synthesis of a complementary strand of RNA from a DNA template. • Enzymes that bond nucleotides together in a chain to make a new RNA molecule. • Messenger RNA (mRNA) • Form of RNA that carries genetic information from the nucleus to the cytoplasm, where ...
Module 7: The Central Dogma
... Transfer RNA (tRNA) • Each amino acid can have several tRNAs, one for each codon varia=on. • Various tRNA synthetases and other enzymes provide a post-‐transla=onal modifica=on that adds the amino ...
... Transfer RNA (tRNA) • Each amino acid can have several tRNAs, one for each codon varia=on. • Various tRNA synthetases and other enzymes provide a post-‐transla=onal modifica=on that adds the amino ...
Gene Control
... needed for all transcription of genes i. GTFs bind each other & RNA Polym. II to form initiation complex ii. Initiation complex binds to control elements near promotor: start transcription ...
... needed for all transcription of genes i. GTFs bind each other & RNA Polym. II to form initiation complex ii. Initiation complex binds to control elements near promotor: start transcription ...
Zoology 145 course
... • Bacteria have a single type of RNA polymerase that synthesizes all RNA molecules. • In contrast, eukaryotes have three RNA polymerases (I, II, and III) in their nuclei. – RNA polymerase II is used for mRNA synthesis. ...
... • Bacteria have a single type of RNA polymerase that synthesizes all RNA molecules. • In contrast, eukaryotes have three RNA polymerases (I, II, and III) in their nuclei. – RNA polymerase II is used for mRNA synthesis. ...
Lecture 6, Exam III Worksheet Answers
... within the protein? Which causes only minimal damage usually? 1. Silent mutation- causes no change within the protein. A change in a base pair may make one codon into another codon that codes for the exact same amino acid as the first one. 2. Missense mutation- usually causes only minimal damage. Th ...
... within the protein? Which causes only minimal damage usually? 1. Silent mutation- causes no change within the protein. A change in a base pair may make one codon into another codon that codes for the exact same amino acid as the first one. 2. Missense mutation- usually causes only minimal damage. Th ...
Immobilization of Membrane Proteins on Beads
... antibody (Figures 1B-D). The receptor-specific binding of these antibodies indicates that bead-attached GPCRs retain their tertiary structure. A cytoplasmic epitope was not accessible to antibody staining, suggesting that Lipoparticle membranes were not compromised during attachment, washing, or sta ...
... antibody (Figures 1B-D). The receptor-specific binding of these antibodies indicates that bead-attached GPCRs retain their tertiary structure. A cytoplasmic epitope was not accessible to antibody staining, suggesting that Lipoparticle membranes were not compromised during attachment, washing, or sta ...
Ch. 17 From Gene to Protein
... Processing of pre-mRNA Each end of a pre-mRNA molecule is modified in a particular way The 5 end receives a modified nucleotide cap The 3 end gets a poly-A tail A modified guanine nucleotide added to the 5 end ...
... Processing of pre-mRNA Each end of a pre-mRNA molecule is modified in a particular way The 5 end receives a modified nucleotide cap The 3 end gets a poly-A tail A modified guanine nucleotide added to the 5 end ...
SR protein

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are ""S"" and ""R"" respectively. SR proteins are 50-300 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS binding domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.SR proteins were discovered in the 1990s in Drosophila and in amphibian oocytes, and later in humans. In general, metazoans appear to have SR proteins and unicellular organisms lack SR proteins.SR proteins are important in constitutive and alternative pre-mRNA splicing, mRNA export, genome stabilization, nonsense-mediated decay, and translation. SR proteins alternatively splice pre-mRNA by preferentially selecting different splice sites on the pre-mRNA strands to create multiple mRNA transcripts from one pre-mRNA transcript. Once splicing is complete the SR protein may or may not remain attached to help shuttle the mRNA strand out of the nucleus. As RNA Polymerase II is transcribing DNA into RNA, SR proteins attach to newly made pre-mRNA to prevent the pre-mRNA from binding to the coding DNA strand to increase genome stabilization. Topoisomerase I and SR proteins also interact to increase genome stabilization. SR proteins can control the concentrations of specific mRNA that is successfully translated into protein by selecting for nonsense-mediated decay codons during alternative splicing. SR proteins can alternatively splice NMD codons into its own mRNA transcript to auto-regulate the concentration of SR proteins. Through the mTOR pathway and interactions with polyribosomes, SR proteins can increase translation of mRNA.Ataxia telangiectasia, neurofibromatosis type 1, several cancers, HIV-1, and spinal muscular atrophy have all been linked to alternative splicing by SR proteins.