Geuvadis Analysis Meeting
... - Quantified 615 datasets based on the Gencode v7 annotation - Sensitivity is a function of sequencing depth ...
... - Quantified 615 datasets based on the Gencode v7 annotation - Sensitivity is a function of sequencing depth ...
CNVs vs. SNPs: Understanding Human Structural Variation in Disease
... Exhaustive analysis of human single nucleotide polymorphisms or SNPs has led to the identification of interesting genetic markers for certain disorders. But these small changes are not the whole picture. Copy number variations or CNVs, which are the gain or lo ...
... Exhaustive analysis of human single nucleotide polymorphisms or SNPs has led to the identification of interesting genetic markers for certain disorders. But these small changes are not the whole picture. Copy number variations or CNVs, which are the gain or lo ...
Agilent Whole Human Genome Oligo Microarray Kit
... Cutting-edge disease and drug discovery research demands up-to-date content. Agilent has taken extra measures to design its whole genome microarray to represent the best possible view of the human genome as we know it today. The vast majority of highly-sensitive 60-mer oligo probes on this microarra ...
... Cutting-edge disease and drug discovery research demands up-to-date content. Agilent has taken extra measures to design its whole genome microarray to represent the best possible view of the human genome as we know it today. The vast majority of highly-sensitive 60-mer oligo probes on this microarra ...
CGH Microarray Solutions for Genome-Wide Genetic Analysis
... The Agilent microarray workflow is streamlined and takes less time to process samples compared with other microarray methods. Agilent offers all of the reagents and instruments needed to process your aCGH data including custom and catalog arrays, labeling kits, hybridization and wash solutions, hybr ...
... The Agilent microarray workflow is streamlined and takes less time to process samples compared with other microarray methods. Agilent offers all of the reagents and instruments needed to process your aCGH data including custom and catalog arrays, labeling kits, hybridization and wash solutions, hybr ...
Analysis of Toxoplasma gondii Repeat Region 529 bp (NCBI Acc
... classically relies on serology and the demonstration of the pathogen in patient samples. Body fluids or tissues may also be inoculated intra peritoneal into mice or used to infect cell cultures in vitro (Homan et al., 2000), but these methods are less effective. Serological diagnosis of active infec ...
... classically relies on serology and the demonstration of the pathogen in patient samples. Body fluids or tissues may also be inoculated intra peritoneal into mice or used to infect cell cultures in vitro (Homan et al., 2000), but these methods are less effective. Serological diagnosis of active infec ...
Document
... Finding SNPs: HapMap Browser 1. HapMap data sets are useful because individual genotype data in deeply sampled populations can be used to determine optimal genotyping strategies (tagSNPs) or perform population genetic analyses (linkage disequilbrium) ...
... Finding SNPs: HapMap Browser 1. HapMap data sets are useful because individual genotype data in deeply sampled populations can be used to determine optimal genotyping strategies (tagSNPs) or perform population genetic analyses (linkage disequilbrium) ...
High-Throughput Analysis of Foodborne Bacterial Genomic DNA
... which a large number of libraries are to be prepared, for example, with a robotic platform such as the Agilent Bravo Automated Liquid Handling Platform. Both methods are well accepted as reference methods in quantifying dsDNA prior to DNA fragmentation and library construction. To demonstrate the eq ...
... which a large number of libraries are to be prepared, for example, with a robotic platform such as the Agilent Bravo Automated Liquid Handling Platform. Both methods are well accepted as reference methods in quantifying dsDNA prior to DNA fragmentation and library construction. To demonstrate the eq ...
Ensembl Introduction
... • Pilot project completed in 2007: 1% of human genome • Discovered promoter elements are on either side of the transcription start site ...
... • Pilot project completed in 2007: 1% of human genome • Discovered promoter elements are on either side of the transcription start site ...
Statistical analysis of simple repeats in the human genome
... repulsion among words one should expect sL o1: In Fig. 5, we compare the results of our analysis on separations among tracts of direct repeats of given length in the whole human genome with the result of the same analysis performed on short repeats of the type poly(XY), coded according to the alphab ...
... repulsion among words one should expect sL o1: In Fig. 5, we compare the results of our analysis on separations among tracts of direct repeats of given length in the whole human genome with the result of the same analysis performed on short repeats of the type poly(XY), coded according to the alphab ...
Sequence Enhancer Information - Garvan Institute of Medical
... In this report we have demonstrated the powerful effects of betaine, dimethyl sulfoxide, and 7-deaza-dGTP in combination on the amplification of three sequences with a high GC content. These molecules have been shown in the past to enhance amplification separately or in combinations of two, such as ...
... In this report we have demonstrated the powerful effects of betaine, dimethyl sulfoxide, and 7-deaza-dGTP in combination on the amplification of three sequences with a high GC content. These molecules have been shown in the past to enhance amplification separately or in combinations of two, such as ...
Full Text
... the assumption that, in general, sequence similarity is a strong indication of orthology among genes or proteins, these methods are typically performed using a reciprocal best BLAST hit (RBBH) approach. While RBBH detection is computationally fast, it has its own drawbacks, such as being affected by ...
... the assumption that, in general, sequence similarity is a strong indication of orthology among genes or proteins, these methods are typically performed using a reciprocal best BLAST hit (RBBH) approach. While RBBH detection is computationally fast, it has its own drawbacks, such as being affected by ...
#2
... converge to u/(u 1 v), where u and v are, respectively, the AT ! GC and GC ! AT substitution rates. This assumption is, however, not valid in vertebrates, where it is known that the rate of mutation of a given base depends on the nature of its neighboring bases, essentially because of the hypermutab ...
... converge to u/(u 1 v), where u and v are, respectively, the AT ! GC and GC ! AT substitution rates. This assumption is, however, not valid in vertebrates, where it is known that the rate of mutation of a given base depends on the nature of its neighboring bases, essentially because of the hypermutab ...
Clinical application of High throughput sequencing (HTS) analysis
... repeats) or Cri-du-Chat syndrome already have a HTS position although at the moment these disorders are more efficiently detected by traditional methods due to technical limitations or high costs of HTS. As long as no changes occur regarding these issues, you will continue to use for these disorders ...
... repeats) or Cri-du-Chat syndrome already have a HTS position although at the moment these disorders are more efficiently detected by traditional methods due to technical limitations or high costs of HTS. As long as no changes occur regarding these issues, you will continue to use for these disorders ...
First genomic insights into members of a candidate bacterial
... microorganisms (Hulshoff Pol et al., 2004; Li et al., 2008; Yamada & Sekiguchi, 2009) and a phylogenetically novel filament was previously reported to be the cause of bulking in an industrial UASB reactor treating sugar manufacturing wastewater (Yamada et al., 2007; Yamada et al., 2011). Small subun ...
... microorganisms (Hulshoff Pol et al., 2004; Li et al., 2008; Yamada & Sekiguchi, 2009) and a phylogenetically novel filament was previously reported to be the cause of bulking in an industrial UASB reactor treating sugar manufacturing wastewater (Yamada et al., 2007; Yamada et al., 2011). Small subun ...
View Poster - Technology Networks
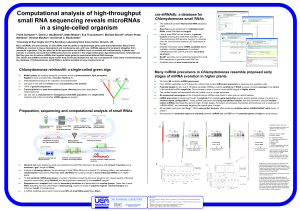
... To find potential miRNA-precursors, a number of windows containing the locus sequence and varying lengths of flanking sequence were folded using RNAfold1 and the significance assessed by the randfold2 program. Sequences were mapped back onto the secondary structures and assembled into overlap groups ...
... To find potential miRNA-precursors, a number of windows containing the locus sequence and varying lengths of flanking sequence were folded using RNAfold1 and the significance assessed by the randfold2 program. Sequences were mapped back onto the secondary structures and assembled into overlap groups ...
CHAPTER 1 LITERATURE SURVEY
... transcribed from plasmids) containing the viral genome directly into cells, as was first demonstrated with Poliovirus (PV; Racaniello & Baltimore 1981). Due to their generally smaller genome sizes compared to DNA viruses, whole RNA virus genomes can be cloned as cDNA and manipulated at will. This ap ...
... transcribed from plasmids) containing the viral genome directly into cells, as was first demonstrated with Poliovirus (PV; Racaniello & Baltimore 1981). Due to their generally smaller genome sizes compared to DNA viruses, whole RNA virus genomes can be cloned as cDNA and manipulated at will. This ap ...
Whole-transcriptome RNAseq analysis from minute amount of total
... alternative splice isoforms and direct measurement of transcript abundance (3). These technologies are greatly accelerating our understanding of the complexity of gene expression, regulation and pathways for mammalian cells. Currently, HT-sequencing technologies have been used for whole-transcriptom ...
... alternative splice isoforms and direct measurement of transcript abundance (3). These technologies are greatly accelerating our understanding of the complexity of gene expression, regulation and pathways for mammalian cells. Currently, HT-sequencing technologies have been used for whole-transcriptom ...
Correlation between sequence divergence and polymorphism
... By visualizing the remaining contigs in Consed v21 [21] and using information regarding reads that span multiple contigs, 63 of the initial contigs were reassembled into nine final contigs with a total length of 147.3 kb and an average single copy coverage depth of 20×. For the ccsA gene, PCR and Sa ...
... By visualizing the remaining contigs in Consed v21 [21] and using information regarding reads that span multiple contigs, 63 of the initial contigs were reassembled into nine final contigs with a total length of 147.3 kb and an average single copy coverage depth of 20×. For the ccsA gene, PCR and Sa ...
Biology Ch. 13
... The Human Genome Project The goal of the Human Genome Project (HGP) was to determine the sequence of the approximately three billion nucleotides that make up human DNA and to identify all of the approximately 20,000–25,000 human genes. ...
... The Human Genome Project The goal of the Human Genome Project (HGP) was to determine the sequence of the approximately three billion nucleotides that make up human DNA and to identify all of the approximately 20,000–25,000 human genes. ...
Syntrophic linkage between predatory Carpediemonas and
... phase and subjected to next-generation sequencing. For this, biomass was harvested from an enrichment culture that had been transferred five times. DNA and RNA extraction was performed as previously described (Smith et al., 2007). For DNA sequencing, a Nextera ...
... phase and subjected to next-generation sequencing. For this, biomass was harvested from an enrichment culture that had been transferred five times. DNA and RNA extraction was performed as previously described (Smith et al., 2007). For DNA sequencing, a Nextera ...
Document
... The assignment of a function to a gene product can be made by a human curator by assessing all of the data (similarities, protein domains, signal peptide etc) This is a labour intensive process and like gene prediction is subjective ...
... The assignment of a function to a gene product can be made by a human curator by assessing all of the data (similarities, protein domains, signal peptide etc) This is a labour intensive process and like gene prediction is subjective ...
human genome research
... sequences of shorter, overlapping fragments. By the 1980s, researchers had started to think seriously about applying such methods to whole genomes. Initial targets were simple organisms such as bacteria, but even these presented formidable challenges: the first sequences of bacterial genomes were on ...
... sequences of shorter, overlapping fragments. By the 1980s, researchers had started to think seriously about applying such methods to whole genomes. Initial targets were simple organisms such as bacteria, but even these presented formidable challenges: the first sequences of bacterial genomes were on ...
ppt
... Main source of new variants within a reproductively isolated species Mutation often ignored because rates assumed to be extremely low relative to magnitude of other effects Accumulation of mutations in population primarily a function of drift and selection PLUS rate of backmutation Mutation rates a ...
... Main source of new variants within a reproductively isolated species Mutation often ignored because rates assumed to be extremely low relative to magnitude of other effects Accumulation of mutations in population primarily a function of drift and selection PLUS rate of backmutation Mutation rates a ...
Supplementary Methods Tables
... The open access tier is publically accessible, and contains data that are considered by TCGA to present a low risk of re-identification of individual participants. The open access data tier does not require user certification for data access. The controlled access tier contains data, including all ...
... The open access tier is publically accessible, and contains data that are considered by TCGA to present a low risk of re-identification of individual participants. The open access data tier does not require user certification for data access. The controlled access tier contains data, including all ...
Whole genome sequencing

Whole genome sequencing (also known as full genome sequencing, complete genome sequencing, or entire genome sequencing) is a laboratory process that determines the complete DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast.Whole genome sequencing should not be confused with DNA profiling, which only determines the likelihood that genetic material came from a particular individual or group, and does not contain additional information on genetic relationships, origin or susceptibility to specific diseases. Also unlike full genome sequencing, SNP genotyping covers less than 0.1% of the genome. Almost all truly complete genomes are of microbes; the term ""full genome"" is thus sometimes used loosely to mean ""greater than 95%"". The remainder of this article focuses on nearly complete human genomes.High-throughput genome sequencing technologies have largely been used as a research tool and are currently being introduced in the clinics. In the future of personalized medicine, whole genome sequence data will be an important tool to guide therapeutic intervention. The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response.