Enzymes used in Genetic Engineering The ability to manipulate
... DNases which act on specific positions or sequences on the DNA are called as restriction endonucleases. The sequences which are recognized by the restriction endonucleases or restriction enzymes (RE) are called as recognition sequences or restriction sites. These sequences are palindromic sequences. ...
... DNases which act on specific positions or sequences on the DNA are called as restriction endonucleases. The sequences which are recognized by the restriction endonucleases or restriction enzymes (RE) are called as recognition sequences or restriction sites. These sequences are palindromic sequences. ...
Pol II, Pol IV and Pol V
... and suf®cient for ±2 and ±1 frameshift mutagenesis induced by the chemical carcinogen N-2-acetylamino¯uorene (AAF), respectively, benzo(a)pyrene (BaP)induced ±1 frameshift mutagenesis requires both Pol IV (dinB) and Pol V. The present work shows that, in response to the vast diversity of existing DN ...
... and suf®cient for ±2 and ±1 frameshift mutagenesis induced by the chemical carcinogen N-2-acetylamino¯uorene (AAF), respectively, benzo(a)pyrene (BaP)induced ±1 frameshift mutagenesis requires both Pol IV (dinB) and Pol V. The present work shows that, in response to the vast diversity of existing DN ...
File
... • DNA duplicates itself prior to cell division. • DNA replication begins with the unwinding of the DNA strands of the double helix. • Each strand is now exposed to a collection of free nucleotides that will be used to recreate the double helix, letter by letter, using base pairing. • Many enzymes an ...
... • DNA duplicates itself prior to cell division. • DNA replication begins with the unwinding of the DNA strands of the double helix. • Each strand is now exposed to a collection of free nucleotides that will be used to recreate the double helix, letter by letter, using base pairing. • Many enzymes an ...
DNA Polymerase I
... Since the parental double helix must rotate 360° to unwind each gyre of the helix, during the semi-conservative replication of the bacterial chromosome, some kind of “swivel” must exist. What do geneticists now know that the required swivel is? a) Topoisomerase ...
... Since the parental double helix must rotate 360° to unwind each gyre of the helix, during the semi-conservative replication of the bacterial chromosome, some kind of “swivel” must exist. What do geneticists now know that the required swivel is? a) Topoisomerase ...
DNA SEQUENCING AND GENE STRUCTURE
... In the first we use a reagent that carries the specificity, but we limit the extent of that reaction - to only one base out of several hundred possible targets in each DNA fragment. This permits the reaction to be used in the domain of greatest specificity: only the very initial stages of a chemical ...
... In the first we use a reagent that carries the specificity, but we limit the extent of that reaction - to only one base out of several hundred possible targets in each DNA fragment. This permits the reaction to be used in the domain of greatest specificity: only the very initial stages of a chemical ...
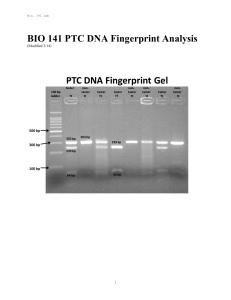
BIO 141 PTC DNA Fingerprint Analysis
... DNA’s structure resembles a twisted ladder called the double helix. DNA in all organisms consists of four bases called guanine, adenine, thymine, and cytosine. The unique order or sequence of these bases in an individual’s cells serves as the blueprint for that individual. Of the approximately 3.3 b ...
... DNA’s structure resembles a twisted ladder called the double helix. DNA in all organisms consists of four bases called guanine, adenine, thymine, and cytosine. The unique order or sequence of these bases in an individual’s cells serves as the blueprint for that individual. Of the approximately 3.3 b ...
Forensic DNA Analysis and the Validation of Applied Biosystems
... (Hallick, 1995). Figure 1 in the Appendix shows the structure of a nucleic acid. The sequence of nucleobases is always listed from the phosphate 5’ end to the hydroxyl 3’ end, to ensure common directionality (Butler, 2010). The DNA molecule is usually found double-stranded in a helix shape, with two ...
... (Hallick, 1995). Figure 1 in the Appendix shows the structure of a nucleic acid. The sequence of nucleobases is always listed from the phosphate 5’ end to the hydroxyl 3’ end, to ensure common directionality (Butler, 2010). The DNA molecule is usually found double-stranded in a helix shape, with two ...
Jeopardy
... CAATTG GTTAAC in a double strand of DNA. If the cut creates two sticky ends that are four bases long, what will one of the exposed sequences (sticky ends) be? ...
... CAATTG GTTAAC in a double strand of DNA. If the cut creates two sticky ends that are four bases long, what will one of the exposed sequences (sticky ends) be? ...
Students
... I know it has been a while, but this is Hope Kelly and I took your AP biology class my junior year of high school at the Career Center. I currently am a sophomore at NC State and have decided to major in human biology with a plan to apply to a graduate program for physical therapy upon graduation. I ...
... I know it has been a while, but this is Hope Kelly and I took your AP biology class my junior year of high school at the Career Center. I currently am a sophomore at NC State and have decided to major in human biology with a plan to apply to a graduate program for physical therapy upon graduation. I ...
Week 12_DNA - Saint Joseph High School
... linking a series of repeating units called nucleotides. • A nucleotide is composed of a sugar, a phosphorouscontaining group, and a nitrogen-containing molecule called a base. • Four types of bases are associated with the DNA structure: adenine (A), guanine (G), cytosine (C), and thymine (T). • As a ...
... linking a series of repeating units called nucleotides. • A nucleotide is composed of a sugar, a phosphorouscontaining group, and a nitrogen-containing molecule called a base. • Four types of bases are associated with the DNA structure: adenine (A), guanine (G), cytosine (C), and thymine (T). • As a ...
12_Lecture_Presentation
... – Recombinant DNA is formed by joining DNA sequences from two different sources – One source contains the gene that will be cloned – Another source is a gene carrier, called a vector ...
... – Recombinant DNA is formed by joining DNA sequences from two different sources – One source contains the gene that will be cloned – Another source is a gene carrier, called a vector ...
CHAPTER 7 DNA Mutation, DNA Repair and Transposable Elements
... i. Different his tester strains are available, to test for base-substitution and frameshift mutations. ii. Liver enzymes (the S9 extract) are mixed with the test chemical to determine whether the liver’s detoxification pathways convert it to a mutagenic form. iii. More revertants in the region of th ...
... i. Different his tester strains are available, to test for base-substitution and frameshift mutations. ii. Liver enzymes (the S9 extract) are mixed with the test chemical to determine whether the liver’s detoxification pathways convert it to a mutagenic form. iii. More revertants in the region of th ...
- Wiley Online Library
... tested, suggesting that this gene may have a conserved function in plants. The protein encoded by this gene is able to correct significantly the sensitivity to the crosslinking agent mitomycin C in ERCC1-deficient Chinese hamster ovary (CHO) cells. These findings suggest that the NER mechanism is co ...
... tested, suggesting that this gene may have a conserved function in plants. The protein encoded by this gene is able to correct significantly the sensitivity to the crosslinking agent mitomycin C in ERCC1-deficient Chinese hamster ovary (CHO) cells. These findings suggest that the NER mechanism is co ...
Recovery response of dividing cells in the thymus of
... S phaseof the division cycleis the most resistantto radiation. These findings are in agreementwith previousobservationson the radiosensitivityand recovery response of other mammalian cell systems after synchrony induced by hydroxyurea(Brown 1,975). In the splenic lymphoid population both the large a ...
... S phaseof the division cycleis the most resistantto radiation. These findings are in agreementwith previousobservationson the radiosensitivityand recovery response of other mammalian cell systems after synchrony induced by hydroxyurea(Brown 1,975). In the splenic lymphoid population both the large a ...
Effect of Supporting Substrates on the Structure of DNA and DNA
... Figure 1 shows typical images of linear DNA molecules sorbed on the modified HOPG surface (DNA was imaged in air). The AFM images are rather stable and are not destroyed on repeated scanning despite the fact that DNA was imaged in contact mode that exerts a more destructive effect of cantilever to t ...
... Figure 1 shows typical images of linear DNA molecules sorbed on the modified HOPG surface (DNA was imaged in air). The AFM images are rather stable and are not destroyed on repeated scanning despite the fact that DNA was imaged in contact mode that exerts a more destructive effect of cantilever to t ...
Tet Proteins Can Convert 5-Methylcytosine to 5
... accumulation of 5fC and 5caC when 5mC is used as a substrate (Fig. 3B, left) strongly suggests that Tet-catalyzed iterative oxidation is likely a kinetically relevant pathway. To determine whether Tet-catalyzed iterative oxidation of 5mC can take place in vivo, we transfected a mammalian expression ...
... accumulation of 5fC and 5caC when 5mC is used as a substrate (Fig. 3B, left) strongly suggests that Tet-catalyzed iterative oxidation is likely a kinetically relevant pathway. To determine whether Tet-catalyzed iterative oxidation of 5mC can take place in vivo, we transfected a mammalian expression ...
Proceedings Template - WORD
... structure provides a backup of all genetic information encoded within double-stranded DNA, i.e., a living organism’s biological information is encoded in these DNA strands. In other words DNA stores the organism’s biological information just as a hard drive stores information on a computer. Several ...
... structure provides a backup of all genetic information encoded within double-stranded DNA, i.e., a living organism’s biological information is encoded in these DNA strands. In other words DNA stores the organism’s biological information just as a hard drive stores information on a computer. Several ...
DNA technology
... • This splicing process can be accomplished by – using restriction enzymes, which cut DNA at specific nucleotide sequences (restriction sites), and – producing pieces of DNA called restriction fragments with “sticky ends” important for joining DNA from different sources. – DNA ligase connects the DN ...
... • This splicing process can be accomplished by – using restriction enzymes, which cut DNA at specific nucleotide sequences (restriction sites), and – producing pieces of DNA called restriction fragments with “sticky ends” important for joining DNA from different sources. – DNA ligase connects the DN ...
SHORT COMMUNICATION A Procedure for Isolating
... from vegetative DNA. No attempt has been made to establish if there are minor differences. The mechanism by which spore lysis is achieved is not fully understood. Could & Hitchins (1963) and many others have shown that treatment with reducing agents at extreme pH values allows spores to become phase ...
... from vegetative DNA. No attempt has been made to establish if there are minor differences. The mechanism by which spore lysis is achieved is not fully understood. Could & Hitchins (1963) and many others have shown that treatment with reducing agents at extreme pH values allows spores to become phase ...
12–1 DNA
... explained how DNA carried information and could be copied. Watson and Crick's model of DNA was a double helix, in which two strands were wound around each other. Slide 30 of 37 Copyright Pearson Prentice Hall ...
... explained how DNA carried information and could be copied. Watson and Crick's model of DNA was a double helix, in which two strands were wound around each other. Slide 30 of 37 Copyright Pearson Prentice Hall ...
DNA
... • The error rate in DNA replication is very low, partly because repair enzymes are able to correct mistakes. • When such mistakes are made and then not corrected, the result is a mutation: a permanent alteration in a cell’s DNA base sequence. ...
... • The error rate in DNA replication is very low, partly because repair enzymes are able to correct mistakes. • When such mistakes are made and then not corrected, the result is a mutation: a permanent alteration in a cell’s DNA base sequence. ...
DNA-dependent protein kinase in nonhomologous end joining: a
... and a DNA terminus. This latter requirement makes the DNAPKCS protein kinase truly DNA dependent. Many targets for the DNA-PKCS kinase have been identified in vitro, including XRCC4, Ku 70/80, Artemis, p53, and even DNA-PKCS itself (autophosphorylation). This plethora of possible targets suggests th ...
... and a DNA terminus. This latter requirement makes the DNAPKCS protein kinase truly DNA dependent. Many targets for the DNA-PKCS kinase have been identified in vitro, including XRCC4, Ku 70/80, Artemis, p53, and even DNA-PKCS itself (autophosphorylation). This plethora of possible targets suggests th ...
DNA repair

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs).The rate of DNA repair is dependent on many factors, including the cell type, the age of the cell, and the extracellular environment. A cell that has accumulated a large amount of DNA damage, or one that no longer effectively repairs damage incurred to its DNA, can enter one of three possible states: an irreversible state of dormancy, known as senescence cell suicide, also known as apoptosis or programmed cell death unregulated cell division, which can lead to the formation of a tumor that is cancerousThe DNA repair ability of a cell is vital to the integrity of its genome and thus to the normal functionality of that organism. Many genes that were initially shown to influence life span have turned out to be involved in DNA damage repair and protection.