Biology
... organism. DNA is very fragile. Chemicals, UV light or other agents can destroy it. Also DNA is unable to leave the nucleus. If it left the nucleus some of the genetic information of the cell would be lost. Instead messenger RNA (mRNA) carries the DNA code from the nucleus into the cytoplasm where th ...
... organism. DNA is very fragile. Chemicals, UV light or other agents can destroy it. Also DNA is unable to leave the nucleus. If it left the nucleus some of the genetic information of the cell would be lost. Instead messenger RNA (mRNA) carries the DNA code from the nucleus into the cytoplasm where th ...
... of a ladder, with hydrogen-bonded base pairs composing the rungs. This ladder is twisted into a helix . The Watson and Crick model also accurately predicted DNA replicates. As the helix unzips, each parental strand serves as the template for the synthesis of a new daughter strand. Through replicatio ...
a copy of the In Search of My Father lab
... boys, the mothers, and the surviving father. Chromosomal DNA, which is present in the nucleus of every living cell, is the genetic material that acts as a blueprint for all of the proteins synthesized by that cell. Unlike mitochondrial DNA, chromosomal DNA is an equal combination of both parents. In ...
... boys, the mothers, and the surviving father. Chromosomal DNA, which is present in the nucleus of every living cell, is the genetic material that acts as a blueprint for all of the proteins synthesized by that cell. Unlike mitochondrial DNA, chromosomal DNA is an equal combination of both parents. In ...
REVIEW - TESADVBiology
... _____ 1. A protein is a polymer consisting of a specific sequence of a. amino acids. c. RNA nucleotides. b. fatty acids. d. DNA nucleotides. _____ 2. The genetic code specifies the correlation between a. a DNA-nucleotide sequence and an RNA-nucleotide sequence. b. an mRNA-nucleotide sequence and a t ...
... _____ 1. A protein is a polymer consisting of a specific sequence of a. amino acids. c. RNA nucleotides. b. fatty acids. d. DNA nucleotides. _____ 2. The genetic code specifies the correlation between a. a DNA-nucleotide sequence and an RNA-nucleotide sequence. b. an mRNA-nucleotide sequence and a t ...
2014 DNA Replication ppt
... In eukaryotic chromosomes, DNA replication occurs at hundreds of places. Replication proceeds in both directions until each chromosome is completely copied. The sites where separation and replication occur are called replication forks. ...
... In eukaryotic chromosomes, DNA replication occurs at hundreds of places. Replication proceeds in both directions until each chromosome is completely copied. The sites where separation and replication occur are called replication forks. ...
DNA Ligase
... 2. How does DNA solve the problem of slow replication on the lagging strand? 3. Code the complementary DNA strand: 3’ T A G C T A A G C T A C 5’ 4. What is the function of telomeres? ...
... 2. How does DNA solve the problem of slow replication on the lagging strand? 3. Code the complementary DNA strand: 3’ T A G C T A A G C T A C 5’ 4. What is the function of telomeres? ...
Human Genetics
... helix is the specific pairing of purines and pyrimidines via hydrogen bonds The complementary base pairs are: - Adenine and guanine - Cytosine and thymine ...
... helix is the specific pairing of purines and pyrimidines via hydrogen bonds The complementary base pairs are: - Adenine and guanine - Cytosine and thymine ...
Chapter 16 DNA
... 2. How does DNA solve the problem of slow replication on the lagging strand? 3. Code the complementary DNA strand: 3’ T A G C T A A G C T A C 5’ 4. What is the function of telomeres? ...
... 2. How does DNA solve the problem of slow replication on the lagging strand? 3. Code the complementary DNA strand: 3’ T A G C T A A G C T A C 5’ 4. What is the function of telomeres? ...
Chapter 3 - About Mrs. Telfort
... What were the variables in Griffith’s experiments? Avery’s Experiments with Nucleic Acids In the 1940s, Oswald Avery wanted to determine whether the transforming agent in Griffith’s experiments was protein, RNA, or DNA. Avery and his colleagues used enzymes to destroy each of these molecules in heat ...
... What were the variables in Griffith’s experiments? Avery’s Experiments with Nucleic Acids In the 1940s, Oswald Avery wanted to determine whether the transforming agent in Griffith’s experiments was protein, RNA, or DNA. Avery and his colleagues used enzymes to destroy each of these molecules in heat ...
Academic Nucleic Acids and Protein Synthesis Test
... 26. Explain the structure and function of DNA 27. Explain the significance of proteins in cell structure and function. 28. Explain the significance of gene transformation. 29. Explain the connection between the DNA code, the mRNA codon, and protein synthesis. ...
... 26. Explain the structure and function of DNA 27. Explain the significance of proteins in cell structure and function. 28. Explain the significance of gene transformation. 29. Explain the connection between the DNA code, the mRNA codon, and protein synthesis. ...
CHAPTER 11D-6
... by a completed Swab Collection Kit form FDLE/FOR-005. The imprinting of the offender’s left and right thumbs, by means of an inked impression, in the spaces indicated on the form shall be completed as well. Inked fingerprint impressions must be legible for fingerprint classification and comparison p ...
... by a completed Swab Collection Kit form FDLE/FOR-005. The imprinting of the offender’s left and right thumbs, by means of an inked impression, in the spaces indicated on the form shall be completed as well. Inked fingerprint impressions must be legible for fingerprint classification and comparison p ...
An essential gene, ESR1, is required for mitotic
... and the RAD50-5 7 genes are involved in meiotic recombination and recombinational repair (3, 19). These observations suggest that the mitotic cell growth, DNA repair and meiotic recombination pathways are closely linked and the genes that are required for them may overlap. The relationships between ...
... and the RAD50-5 7 genes are involved in meiotic recombination and recombinational repair (3, 19). These observations suggest that the mitotic cell growth, DNA repair and meiotic recombination pathways are closely linked and the genes that are required for them may overlap. The relationships between ...
DNA.ppt
... • are the building blocks of your cells • can speed up reactions when they act as enzymes • perform important functions (e.g. hemoglobin – transports oxygen in your blood) • consist of amino acids – All proteins in your body are made by the same 20 amino acids. – What separates one protein from anot ...
... • are the building blocks of your cells • can speed up reactions when they act as enzymes • perform important functions (e.g. hemoglobin – transports oxygen in your blood) • consist of amino acids – All proteins in your body are made by the same 20 amino acids. – What separates one protein from anot ...
DNA (Deoxyribonucleic Acid)
... is a single circular molecule that contains nearly all the cell’s genetic information. ...
... is a single circular molecule that contains nearly all the cell’s genetic information. ...
Document
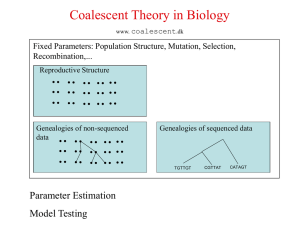
... Diploid Model with Recombination An individual is made by: 1. The paternal chromosome is taken by picking random father. 2. Making that father’s chromosomes recombine to create the individuals paternal chromosome. Similarly for maternal chromosome. ...
... Diploid Model with Recombination An individual is made by: 1. The paternal chromosome is taken by picking random father. 2. Making that father’s chromosomes recombine to create the individuals paternal chromosome. Similarly for maternal chromosome. ...
MCDB 1041: Using DNA To manipulate DNA in the laboratory, one
... 5. The gene for sickle cell anemia is 23 KB in length. You have isolated this fragment from human cells with PCR. You also know that this gene has two sites for the same restriction enzyme, located at 10 KB and at 18 KB in the sequence. a. How many pieces of DNA will be generated by cutting this pie ...
... 5. The gene for sickle cell anemia is 23 KB in length. You have isolated this fragment from human cells with PCR. You also know that this gene has two sites for the same restriction enzyme, located at 10 KB and at 18 KB in the sequence. a. How many pieces of DNA will be generated by cutting this pie ...
Restriction Enzyme Digestion
... • Usually, organisms that make restriction enzymes also make a companion modification enzyme (DNA methyltransferasemethylase) that protects their own DNA from cleavage. • These enzymes recognize the same DNA sequence as the restriction enzyme they accompany, but instead of cleaving the sequence, the ...
... • Usually, organisms that make restriction enzymes also make a companion modification enzyme (DNA methyltransferasemethylase) that protects their own DNA from cleavage. • These enzymes recognize the same DNA sequence as the restriction enzyme they accompany, but instead of cleaving the sequence, the ...
A - sandsbiochem
... d. It contains the nitrogenous base uracil. 29. Individuals with one form of lactose intolerance do not produce the enzyme lactase because the gene coding for the production of lactase is shut off in their cells. This means that which of the following processes does not occur for the gene? a. Hydrog ...
... d. It contains the nitrogenous base uracil. 29. Individuals with one form of lactose intolerance do not produce the enzyme lactase because the gene coding for the production of lactase is shut off in their cells. This means that which of the following processes does not occur for the gene? a. Hydrog ...
Molecular Bio Questions1
... sequences buried in the center of the molecule, so that proteins can interact with the bases without the strands being separated. a. Cohesive ends d. Attractions b. Grooves e. Repulsions c. Nicks ____ 4. A nucleoid gently released from E. coli appears as 30–100 tightly wound loops, each defined by a ...
... sequences buried in the center of the molecule, so that proteins can interact with the bases without the strands being separated. a. Cohesive ends d. Attractions b. Grooves e. Repulsions c. Nicks ____ 4. A nucleoid gently released from E. coli appears as 30–100 tightly wound loops, each defined by a ...
Questions 4
... sequences buried in the center of the molecule, so that proteins can interact with the bases without the strands being separated. a. Cohesive ends d. Attractions b. Grooves e. Repulsions c. Nicks ____ 4. A nucleoid gently released from E. coli appears as 30–100 tightly wound loops, each defined by a ...
... sequences buried in the center of the molecule, so that proteins can interact with the bases without the strands being separated. a. Cohesive ends d. Attractions b. Grooves e. Repulsions c. Nicks ____ 4. A nucleoid gently released from E. coli appears as 30–100 tightly wound loops, each defined by a ...
PPT File
... one pentose and the 5’-OH of the next • the pentose unit is -D-ribose (it is 2-deoxy-D-ribose in DNA) • the pyrimidine bases are uracil and cytosine (they are thymine and cytosine in DNA) • in general, RNA is single stranded (DNA is double stranded) ...
... one pentose and the 5’-OH of the next • the pentose unit is -D-ribose (it is 2-deoxy-D-ribose in DNA) • the pyrimidine bases are uracil and cytosine (they are thymine and cytosine in DNA) • in general, RNA is single stranded (DNA is double stranded) ...
XXII – DNA cloning and sequencing Outline
... a) A heterogenous group of DNA segments inserted into a plasmid (vector; ex. pUC18) using EcoRΙ (makes sticky ends) and DNA ligase. A vector joined to a desired DNA fragment is a recombinant DNA molecule. Plasmid and DNA fragment must be exposed to the same restriction enzyme. b) Recombinant DNA mol ...
... a) A heterogenous group of DNA segments inserted into a plasmid (vector; ex. pUC18) using EcoRΙ (makes sticky ends) and DNA ligase. A vector joined to a desired DNA fragment is a recombinant DNA molecule. Plasmid and DNA fragment must be exposed to the same restriction enzyme. b) Recombinant DNA mol ...
Homologous recombination
Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks. Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.Although homologous recombination varies widely among different organisms and cell types, most forms involve the same basic steps. After a double-strand break occurs, sections of DNA around the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule then ""invades"" a similar or identical DNA molecule that is not broken. After strand invasion, the further sequence of events may follow either of two main pathways discussed below (see Models); the DSBR (double-strand break repair) pathway or the SDSA (synthesis-dependent strand annealing) pathway. Homologous recombination that occurs during DNA repair tends to result in non-crossover products, in effect restoring the damaged DNA molecule as it existed before the double-strand break.Homologous recombination is conserved across all three domains of life as well as viruses, suggesting that it is a nearly universal biological mechanism. The discovery of genes for homologous recombination in protists—a diverse group of eukaryotic microorganisms—has been interpreted as evidence that meiosis emerged early in the evolution of eukaryotes. Since their dysfunction has been strongly associated with increased susceptibility to several types of cancer, the proteins that facilitate homologous recombination are topics of active research. Homologous recombination is also used in gene targeting, a technique for introducing genetic changes into target organisms. For their development of this technique, Mario Capecchi, Martin Evans and Oliver Smithies were awarded the 2007 Nobel Prize for Physiology or Medicine.