Chapter 5 - FIU Faculty Websites
... mammal genome ~ 3-4 billion base pairs of DNA one gene ~ 10 kbp It would be unpractical to isolate 1 gene Preparation of genomic DNA library Isolating total DNA – Aplying shotgun cloning Digesting total DNA into fragments of suitable size (the fragments do not corresponds to the individual genes) In ...
... mammal genome ~ 3-4 billion base pairs of DNA one gene ~ 10 kbp It would be unpractical to isolate 1 gene Preparation of genomic DNA library Isolating total DNA – Aplying shotgun cloning Digesting total DNA into fragments of suitable size (the fragments do not corresponds to the individual genes) In ...
Solid Tumour Section Uterus: Carcinoma of the cervix in Oncology and Haematology
... Chromosome 3: additional material on 3q has been shown by comparative genomic hybridization (CGH) in 90% of carcinomas and this gain may occur at the point of transition from severe dysplasia to invasive carcinoma; loss of heterozygosity (LOH) studies indicate that there are two regions on 3p where ...
... Chromosome 3: additional material on 3q has been shown by comparative genomic hybridization (CGH) in 90% of carcinomas and this gain may occur at the point of transition from severe dysplasia to invasive carcinoma; loss of heterozygosity (LOH) studies indicate that there are two regions on 3p where ...
Chap 4 Chemical Synhesis Sequencing and Amplification of DNA
... Mismatch of primers (to generate wrong, shorter strand insufficient dNTP for correct synthesis) ...
... Mismatch of primers (to generate wrong, shorter strand insufficient dNTP for correct synthesis) ...
Unit 1 content check list
... Give examples of each main form of protein shape (fibrous, globular, conjugated) Explain the need for cellular differentiation Describe how plants (meristems) and animals (stem cells) form specialised cells Describe the difference between; pleuripotent, totipotent and differentiated Give examples of ...
... Give examples of each main form of protein shape (fibrous, globular, conjugated) Explain the need for cellular differentiation Describe how plants (meristems) and animals (stem cells) form specialised cells Describe the difference between; pleuripotent, totipotent and differentiated Give examples of ...
Procedure - DNA Interactive
... cycler is set and have all experimenters set up PCR reactions coordinately. Add primer/loading dye mix to all reaction tubes, then add each student template, and begin ...
... cycler is set and have all experimenters set up PCR reactions coordinately. Add primer/loading dye mix to all reaction tubes, then add each student template, and begin ...
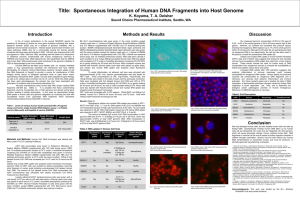
Title: Spontaneous Integration of Human DNA Fragments into Host
... up exogenous DNA in our experiments, the cell line has been used in the past as a model for spontaneous DNA uptake (8). Cellular and nuclear DNA uptake in human foreskin fibroblast (HFF1) cells and in NCCIT cells suggests that embryonic and neonatal cell are more susceptible to DNA uptake than cells ...
... up exogenous DNA in our experiments, the cell line has been used in the past as a model for spontaneous DNA uptake (8). Cellular and nuclear DNA uptake in human foreskin fibroblast (HFF1) cells and in NCCIT cells suggests that embryonic and neonatal cell are more susceptible to DNA uptake than cells ...
Section 20.1
... • Vectors are carrier DNA molecules that can replicate cloned DNA fragments in a host cell • Vectors must be able to replicate independently and should have several restriction enzyme sites to allow insertion of a DNA fragment • Vectors should carry a selectable gene marker to distinguish host cells ...
... • Vectors are carrier DNA molecules that can replicate cloned DNA fragments in a host cell • Vectors must be able to replicate independently and should have several restriction enzyme sites to allow insertion of a DNA fragment • Vectors should carry a selectable gene marker to distinguish host cells ...
Lesson Plans Teacher: Robinson Dates: 2/6
... I can recognize autosomal chromosomes, sexual chromosomes, trisomy chromosomes when reviewing a karyotype. I can explain simple inheritance patterns using pedigrees. Use Punnett's squares to produce BOTH the F1 and F2 generations from a breeding between PPGG x ppgg. (Note to students: F2 are produce ...
... I can recognize autosomal chromosomes, sexual chromosomes, trisomy chromosomes when reviewing a karyotype. I can explain simple inheritance patterns using pedigrees. Use Punnett's squares to produce BOTH the F1 and F2 generations from a breeding between PPGG x ppgg. (Note to students: F2 are produce ...
GENETICS: BIOLOGY HSA REVIEW
... b 1. If a homozygous German Shepherd with brown eyes is bred with a heterozygous German Shepherd with blue eyes, is it possible to have puppies with brown eyes? a. Yes, and all potential offspring will be carriers of the allele for blue eyes. b. Yes; 50% of the offspring would be heterozygous for bl ...
... b 1. If a homozygous German Shepherd with brown eyes is bred with a heterozygous German Shepherd with blue eyes, is it possible to have puppies with brown eyes? a. Yes, and all potential offspring will be carriers of the allele for blue eyes. b. Yes; 50% of the offspring would be heterozygous for bl ...
Word Work File L_2.tmp
... Eukaryotic chromosomal DNA molecules have special nucleotide sequences called telomeres at their ends. These end caps of repetitive DNA are called telomeres. Telomeres do not contain genes. They are made of multiple repetitions of a nucleotide sequence, e. g. TTAGGG is the repetitive unit in humans ...
... Eukaryotic chromosomal DNA molecules have special nucleotide sequences called telomeres at their ends. These end caps of repetitive DNA are called telomeres. Telomeres do not contain genes. They are made of multiple repetitions of a nucleotide sequence, e. g. TTAGGG is the repetitive unit in humans ...
LECTURE 1 Human Chromosomes Human Karyotype
... The packaging of DNA into chromosomes involves several orders of DNA coiling and folding. The normal human karyotype is made up of 46 chromosomes consisting of 22 pairs of autosomes and a pair of sex chromosomes, XX in the female, and XY in the male. Each chromosome consists of a short (p) and ...
... The packaging of DNA into chromosomes involves several orders of DNA coiling and folding. The normal human karyotype is made up of 46 chromosomes consisting of 22 pairs of autosomes and a pair of sex chromosomes, XX in the female, and XY in the male. Each chromosome consists of a short (p) and ...
Application of Molecular Techniques to Improved Detection of
... reaction, resulting in an exponential increase in copy number of a desired nucleotide sequence within hours. The results of this reaction can be easily visualized by electrophoresis in the presence of ethidium bromide, a dye that is selective for nucleic acids. Several applications of the PCR have p ...
... reaction, resulting in an exponential increase in copy number of a desired nucleotide sequence within hours. The results of this reaction can be easily visualized by electrophoresis in the presence of ethidium bromide, a dye that is selective for nucleic acids. Several applications of the PCR have p ...
f^*Co*e -z`
... grow in presence of tetra. This is insertional inactivation. cells are first grown ln media containing tetra and a replica is made on a plate containing amp in the medium. which grow on master plate but fail to grow on the replica plate contain the "t]t: ...
... grow in presence of tetra. This is insertional inactivation. cells are first grown ln media containing tetra and a replica is made on a plate containing amp in the medium. which grow on master plate but fail to grow on the replica plate contain the "t]t: ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.