* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lab 11
Survey
Document related concepts
Gaseous signaling molecules wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Biosynthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
15-Hydroxyeicosatetraenoic acid wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Butyric acid wikipedia , lookup
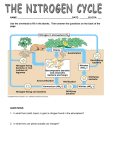
Nitrogen cycle wikipedia , lookup
Metalloprotein wikipedia , lookup
Transcript
Lab 11 Goals and Objectives: Exercise 39: Oxidation and Fermentation Tests Read results: some tubes will require additional reagents Do controls first so you have something to compare to! Exercise 40: Hydrolytic and Degradative Reactions Set up according to Fig 40.1 except both of your two unknowns well separated on each type of plate and control on a different plate (one control plate per pair) ***Save streak plates of unknowns for use next class*** Durham Sugar Tube Fermentation (Glucose, Lactose, Mannitol) Contains: single carbohydrate peptone broth with durham tube for gas collection, Phenol red pH indicator: alkaline pH = red, acidic pH = yellow Discriminates the ability to ferment a single carbohydrate (glucose, lactose, or mannitol) into acid products (e.g. pyruvic acid) or acid plus gas Results: Red = inert, negative for fermentation of specified carbohydrate Yellow = positive for fermentation of carbohydrate to acid products Yellow with bubble = positive for fermentation of carbohydrate to acid + gas Control: Escherichia coli Acid plus gas Negative Acid MR-VP Medium: Methyl Red Test Contains: peptone, glucose, and buffer (buffer will neutralize weak acids so only strong stable acids will be detected by methyl red) Additional reagents added: methyl red pH indicator: acid pH = red, neutral or alkaline pH = yellow Distinguishes ability to catabolize glucose into stable mixed acids (lactic, acetic, and formic acids) in the mixed acid pathway Results: Red = positive for mixed acid formation Yellow = negative for mixed acid formation + Control E.coli _ MR-VP Medium: Voges-Proskauer Test Contains: peptone and glucose Additional reagents added: Barritt’s A (alpha napthol) and Barritt’s B (KOH) (will react with acetoin to produce a red product, alone produce a copper colored product) Distinguishes the ability to catabolize glucose into the neutral end product butanediol (the oxidized product is acetoin) in the butylene glycol pathway Results: Red = positive for acetoin and thus for 2,3-butanediol production Yellow/Orange = no acetoin, negative for 2,3-butanediol production + Control Enterobacter cloacae _ + _ Nitrate Reduction Broth Contains: beef extract, peptone, KNO3 as nitrate source, durham tube for gas collection Additional reagents added: sulfanilic acid (reagent A), dimethylalpha-naphthylamine (reagent B), (together form a complex with nitrite creating a red product), zinc (reduces nitrate to nitrite allowing reaction with reagent A and B) Discriminates organisms that can produce nitrate reductases to utilize nitrate as a final electron acceptor resulting in the production of either nitrite (partial reduction) or to NH4, N2O or N2 gas (complete reduction). Results: Red with reagents A and B = positive for nitrate to nitrite reduction Clear with/wo gas = positive for complete reduction of nitrate to nitrogen gas (or nongaseous N2O or NH4) Red only after zinc = negative for nitrate reduction, negative for nitrate reductases Controls: Pseudomonas aeruginosa and E. coli Anaerobic respiration - Nitrate reduction Electron acceptor Products NO3– NO2–, N2 + H2O SO4– H2S + H2O CO32 – CH4 + H2O NO3- NO2nitrate nitrite NH4 ammonium NO N2O nitric oxide Nitrous oxide N2 molecular nitrogen sulfanilic acid (reagent A) + dimethylalpha-naphthylamine (reagent B) 1. A + B + nitrite = red 2. Zinc converts nitrate to nitrite Nitrate to nitrite add zinc to negative tubes No reduction Complete reduction Simmon’s Citrate Agar Contains: citrate as sole carbon source, ammonium salts as sole nitrogen source, bromthymol blue pH indicator: neutral pH = green, alkaline = prussian blue Discriminates organisms that can produce citrase to metabolize citrate into oxaloacetate and pyruvate. These organisms are forced to utilize ammonium salts as the nitrogen source producing alkaline ammonia waste. Results: Prussian blue slant and or butt = positive for _ + citrase production Green = negative for citrase production CitrateOxaloacetate Pyruvate + ammonium salt Fermentation Alkaline pH Control: Enterobacter cloacae Oxidase Test Discriminates organisms that can produce cytochrome oxidase which catalyzes the transfer of electrons from reduced cytochrome c in the electron transport chain to molecular oxygen. Test uses NNNN-tetramethyl-p-phenylenediamine (Oxidase Reagent) as an artificial electron acceptor: when oxidized it is colorless, when reduced it turns purple *Look for color change on the bacteria, not on the cotton swab! (The reagent will turn light purple when exposed to oxygen in the air) Catalase Test Discriminates aerobic organisms that produce catalase to degrade hydrogen peroxide into water and oxygen + _ http://ftp.ccccd.edu/dcain/CCCCD%20Micro/Catalase.jpg 12 Possible Unknowns Gram Negative Gram Positive Bacillus subtilis Catalase + Gelatinase - Gelatinase + Catalase - Pseudomonas aeruginosa Gelatinase + Gelatinase - Exercise 40: Hydrolytic and Degradative Reactions Set up according to Fig 40.1 except both of your two unknowns well separated on each type of plate and control on a different plate (one control plate per pair) ***Save streak plates of unknowns for use next class*** Fig. 40.1 Unknowns Separate Plates! broth Control broth • Each pair needs: 3 Starch plates 3 Skim milk plates 3 Spirit blue plates 5 Urea broths (replaces urea slant) 5 Phenylalanine slants 5 Tryptone broths • One set of controls per pair using broth cultures: • • • • Bacillus subtilis Staphylococcus aureus Escherichia coli Proteus vulgaris Title Identification of an Unknown Bacterial Culture Laboratory Report • Introduction ( Why the experiment is important? ) – State a hypothesis (an “if then” statement, may require multiple sentences) that is clear and appropriately addresses the purpose of this laboratory exercise. • Materials and Methods (How each of the assay was performed?) – Describe each essential assay separately in a separate paragraph. – Media used to grow the organism and list any reagents or indicators – Do not include the enzymatic reactions here: save those for the Discussion • Results (What are the results observed?) – Include only the key assays, not every test that was performed. In cases where multiple assays were employed to determine one characteristic, only one type of assay need be presented. • Discussion ( Discussion of the assays and interpretation of the results) – Discuss only the assays for which you presented results in the table in the – List these enzymes, substrates, products, and color change • Conclusions (Interpretation of your collective results) – Analyze the results and determine the identification of the unknown based on the collective results – follow the dichotomous key. – Approve or disapprove the hypothesis • Literature Cited – List here all source material cited in the laboratory report. Use proper CSE format for scientific publications including listing the sources in this section in alphabetical order by the first authors last name