* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mendel`s 2 nd Law – Independent Assortment
Human genetic variation wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
X-inactivation wikipedia , lookup
Gene therapy wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Public health genomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genomic imprinting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
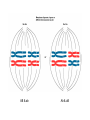
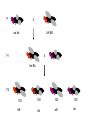
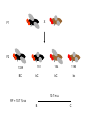
Mendel’s 1st Law - Segregation Each individual has two alleles (copies) of every gene. These alleles separate during sperm/egg cell formation Mendel’s 2nd Law – Independent Assortment During gamete formation, different pairs of alleles segregate independently of each other Potential gametes AaBb AB 1/4 Ab 1/4 aB 1/4 ab 1/4 AB & ab Ab & aB P X aa bb AA BB X F1 Aa Bb F2 100 AB 100 100 100 Ab aB ab P X BB CC bb cc X F1 Bb Cc F2 1339 151 154 1195 BC bC Bc bc Gametes All parental 50% parental 50% recombinant X F1 F2 1339 BC 151 bC 154 1195 bC bc 10.7 m.u. RF = 10.7 % so B C DCO lead to underestimation of genetic map distances Solution is to use three point cross A female Drosophila is heterozygous at three autosomal linked gene loci: e+ h+ c+ brown body hairless legs straight wings e h c ebony body hairy legs curvy wings In order to map these genes, this heterozygote is crossed to a homozygous male recessive fly and the following phenotypes of progeny were obtained: ehc e+ h+ c+ e+ h c e h+ c+ e h c+ e h+ c e+ h+ c e+ h c+ 356 347 2 3 42 19 53 22 844 Why didn’t we do the cross the other way round i.e. with a heterozygous male? Which is the middle gene? Calculate the map distances between the genes. Is there any evidence of interference? (1) (2) (4) (2)