* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download SARS Outbreaks in Ontario, Hong Kong and Singapore: the role of
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Human genetic variation wikipedia , lookup
Gene expression programming wikipedia , lookup
Behavioural genetics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Medical genetics wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
Heritability of IQ wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genetic drift wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Population genetics wikipedia , lookup
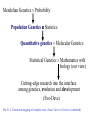
Basic Genetics (1) Mendelian genetics How does a gene transmit from a parent to its progeny (individual)? (2) Population genetics How is a gene segregating in a population (a group of individuals)? (3) Quantitative genetics How is gene segregation related with the phenotype of a character? (4) Molecular genetics What is the molecular basis of gene segregation and transmission? (5) Developmental genetics (6) Epigenetics (genetic imprinting) Genomic Imprinting The callipygous animals 1 and 3 compared to normal animals 2 and 4 (Cockett et al. Science 273: 236-238, 1996) Mendelian Genetics Probability Population Genetics Statistics Quantitative genetics Molecular Genetics Statistical Genetics Mathematics with biology (our view) Cutting-edge research into the interface among genetics, evolution and development (Evo-Deve) Wu, R. L. Functional mapping of complex traits. Nature Reviews Genetics (submitted). Mendel’s Laws Mendel’s first law • There is a gene with two alleles on a chromosome location (locus) • These alleles segregate during the formation of the reproductive cells, thus passing into different gametes Diploid Gene A A| a | Centromere A| Probability A pair of chromosomes ½ Gamete a| ½ Gamete Mendel’s second law • There are two or more pairs of genes on different chromosomes • They segregate independently (partially correct) Diploid A|a|, B|b| Probability A|, B| A|, b| a|, B| a|, b| ¼ ¼ ¼ ¼ Four two-gene gametes What about three genes? Linkage (exception to Mendel’s second law) • There are two or more pairs of genes located on the same chromosome • They can be linked or associated (the degree of association is described by the recombination fraction) High linkage Low linkage A B A B How the linkage occurs? – consider two genes A and B 1 2 3 A a A Aa a B b B B b b A 4 a A a A a A a B B b b B B b b Stage 1: A pair of chromosomes, one from the father and the other from the mother Stage 2: Each chromosome is divided into two sister chromatids Stage 3: Non-sister chromatids crossover Stage 4: Meiosis generates four gametes AB, aB, Ab and ab – Nonrecombinants (AB and ab) and Recombinants (aB and Ab) How to measure the linkage? – based on a design Parents Gamete AABB AB aabb ab × F1 Gamete AaBb AB Ab aB × aabb ab ab Backcross Observations AaBb n1 Aabb n2 aaBb n3 aabb n4 Gamete type Non-recom/ Parental Recom/ Non-parental Recom/ Non-parental Non-recom/ Parental Define the proportion of the recombinant gametes over the total gametes as the recombination fraction (r) between two genes A and B r = (n2+n3)/(n1+n2+n3+n4) Several concepts Genotype and Phenotype • Locus (loci), chromosomal location of a gene • Allele (A, a), a copy of gene • Dominant allele, one allele whose expression inhibits the expression of its alternative allele • Recessive allele (relative to dominant allele) • Dominant gene (AA and Aa are not distinguishable, denoted by A_) • Codominant gene (AA, Aa and aa are mutually distinguishable) • Genotype (AA, Aa or aa) • Homozygote (AA or aa) • Heterozygote (Aa) • Phenotype: trait value Chromosome and Meiosis • Chromosome: Rod-shaped structure made of DNA • Diploid (2n): An organism or cell having two sets of chromosomes or twice the haploid number • Haploid (n): An organism or cell having only one complete set of chromosomes • Gamete: Reproductive cells involved in fertilization. The ovum is the female gamete; the spermatozoon is the male gamete. • Meiosis: A process for cell division from diploid to haploid (2n n) (two biological advantages: maintaining chromosome number unchanged and crossing over between different genes) • Crossover: The interchange of sections between pairing homologous chromosomes during meiosis • Recombination, recombinant, recombination fraction (rate, frequency): The natural formation in offspring of genetic combinations not present in parents, by the processes of crossing over or independent assortment. Molecular markers • Genetic markers are DNA sequence polymorphisms that show Mendelian inheritance • Marker types - Restriction fragment length polymorphism (RFLP) - Amplified fragment length polymorphism (AFLP) - Simple sequence repeat (SSR) - Single nucleotide polymorphism (SNP) Population Genetics • Different copies of a gene are called alleles; for example A and a at gene A; • These alleles form three genotypes, AA, Aa and aa; • The allele (or gene) frequency of an allele is defined as the proportion of this allele among a group of individuals; • Accordingly, the genotype frequency is the proportion of a genotype among a group of individuals Calculations of allele frequencies and genotype frequencies Genotypes Counts AA 224 Aa 64 aa 6 Total 294 Estimates genotype frequencies PAA = 224/294 = 0.762 PAa = 64/294 = 0.218 Paa = 6/294 = 0.020 PAA + PAa + Paa = 1 Allele frequencies pA = (2214+64)/(2294)=0.871, pa = (26+64)/(2294)=0.129, pA + pa = 0.871 + 0.129 = 1 Expected genotype frequencies AA pA2 = 0.8712 = 0.769 Aa 2pApa = 2 0.871 0.129 = 0.224 Aa pa2 = 0.1292 = 0.017 Genotypes AA Aa aa Total Counts nAA nAa naa n Estimates of genotype freq. PAA = nAA/n PAa = nAa/n Paa = naa/n PAA + PAa + Paa = 1 Allele frequencies pA = (2nAA + nAa)/2n pa = (2naa + nAa)/2n Standard error of the estimate of the allele frequency Var(pA) = pA(1 - pA)/2n The Hardy-Weinberg Law • In the Hardy-Weinberg equilibrium (HWE), the relative frequencies of the genotypes will remain unchanged from generation to generation; • As long as a population is randomly mating, the population can reach HWE from the second generation; • The deviation from HWE, called Hardy-Weinberg disequilibrium (HWD), results from many factors, such as selection, mutation, admixture and population structure… Mendelian inheritance at the individual level (1) Make a cross between two individual parents (2) Consider one gene (A) with two alleles A and a AA, Aa, aa Thus, we have a total of nine possible cross combinations: 1. 2. 3. 4. 5. 6. 7. 8. 9. Cross AA AA AA Aa AA aa Aa AA Aa Aa Aa aa aa AA aa Aa aa aa Mendelian segregation ratio AA ½AA + ½Aa Aa ½AA + ½Aa ¼AA + ½Aa + ¼aa ½Aa + ½aa Aa ½Aa + ½aa aa Mendelian inheritance at the population level • A population, a group of individuals, may contain all these nine combinations, weighted by the mating frequencies. • Genotype frequencies: AA, PAA; Aa, PAa; aa, Paa Cross 1. 2. 3. 4. 5. 6. 7. 8. 9. AA AA AA Aa AA aa Aa AA Aa Aa Aa aa aa AA aa Aa aa aa Mating freq. (t) PAA(t)PAA(t) PAA(t)PAa(t) PAA(t)Paa(t) PAa(t)PAA(t) PAa(t)PAa(t) PAa(t)Paa(t) Paa(t)PAA(t) Paa(t)PAa(t) Paa(t)Paa(t) Mendelian segreg. ratio (t+1) AA Aa aa 1 0 0 ½ ½ 0 0 1 0 ½ ½ 0 ¼ ½ ¼ 0 ½ ½ 0 1 0 0 ½ ½ 0 0 1 PAA(t+1) = 1[PAA(t)]2 + ½ 2[PAA(t)PAa(t)] + ¼[PAa(t)]2 = [PAA(t) + ½PAa(t)]2 Similarly, we have Paa(t+1) = [Paa(t) + ½PAa(t)]2 PAa(t+1) = 2[PAA(t) + ½PAa(t)][Paa(t) + ½PAa(t)] Therefore, we have [PAa(t+1)]2 = 4PAA(t+1)Paa(t+1) Furthermore, if random mating continues, we have PAA(t+2) = [PAA(t+1) + ½PAa(t+1)]2 = PAA(t+1) PAa(t+2) = 2[PAA(t+1) + ½PAa(t+1)][Paa(t+1) + ½PAa(t+1)] = PAa(t+1) Paa(t+2) = [Paa(t+1) + ½PAa(t+1)]2 = Paa(t+1) Concluding remarks A population with [PAa(t+1)]2 = 4PAA(t+1)Paa(t+1) is said to be in Hardy-Weinberg equilibrium (HWE). The HWE population has the following properties: (1) Genotype (and allele) frequencies are constant from generation to generation, (2) Genotype frequencies = the product of the allele frequencies, i.e., PAA = pA2, PAa = 2pApa, Paa = pa2 For a population at Hardy-Weinberg disequilibrium (HWD), we have • PAA = pA2 + D • PAa = 2pApa – 2D • Paa = pa2 + D The magnitude of D determines the degree of HWD. • D = 0 means that there is no HWD. • D has a range of max(-pA2 , -pa2) D pApa Chi-square test for HWE • Whether or not the population deviates from HWE at a particular locus can be tested using a chi-square test. • If the population deviates from HWE (i.e., Hardy-Weinberg disequilibrium, HWD), this implies that the population is not randomly mating. Many evolutionary forces, such as mutation, genetic drift and population structure, may operate. Example 1 AA Obs 224 Exp n(pA2) = 222.9 Aa 64 n(2pApa) = 66.2 aa 6 n(pa2) = 4.9 Total 294 294 Test statistics x2 = (obs – exp)2 /exp = (224-222.9)2/222.9 + (64-66.2)2/66.2 + (6-4.9)2/4.9 = 0.32 is less than x2df=1 ( = 0.05) = 3.841 Therefore, the population does not deviate from HWE at this locus. Why the degree of freedom = 1? Degree of freedom = the number of parameters contained in the alternative hypothesis – the number of parameters contained in the null hypothesis. In this case, df = 2 (pA or pa and D) – 1 (pA or pa) = 1 Example 2 Obs Exp AA 234 n(pA2) = 230.1 Aa 36 n(2pApa) = 43.8 aa 6 n(pa2) = 2.1 Total 276 276 Test statistics x2 = (obs – exp)2/exp = (234-230.1)2/230.1 + (3643.8)2/43.8 + (6-2.1)2/2.1 = 8.8 is greater than x2df=1 ( = 0.05) = 3.841 Therefore, the population deviates from HWE at this locus. Linkage disequilibrium • Consider two loci, A and B, with alleles A, a and B, b, respectively, in a population • Assume that the population is at HWE • If the population is at Hardy-Weinberg equilibrium, we have Gene A AA: PAA = pA2 Aa: PAa = 2pApa Aa: Paa = pa2 PAA+PAa+Paa = 1 pA + pa = 1 Gene B BB: PBB = pB2 Bb: PBb = 2pBpb bb: Pbb = pb2 PBB+PBb+Pbb=1 pB + p b = 1 But the population is at Linkage Disequilibrium (for a pair of loci). Then we have • Two-gene haplotype AB: pAB = pApB + DAB • Two-gene haplotype Ab: pAb = pApb + DAb • Two-gene haplotype aB: paB = papB + DaB • Two-gene haplotype ab: pab = papb + Dab pAB+pAb+paB+pab = 1 Dij is the coefficient of linkage disequilibrium (LD) between the two genes in the population. The magnitude of D reflects the degree of LD. The larger D, the stronger LD. pA = pAB+pAb = pApB + DAB + pApb + DAb = pA+DAB+DAb pB = pAB+paB = pB+DAB+DaB pb = pAb+pab = pb+DaB+Dab DAB = -DAb DAB = -DaB Dab = -DaB Finally, we have DAB = -DAb = -DaB = Dab = D. Re-write four two-gene haplotypes • AB: pAB = pApB + D • Ab: pAb = pApb – D • aB: paB = papB – D • ab: pab = papb + D D = pABpab - pAbpaB D = 0 the population is at the linkage equilibrium How does D transmit from one generation (1) to the next (2)? D(2) = (1-r)1 D(1) … D(t+1) = (1-r)t D(1) t, D(t+1) r Proof to D(t+1) = (1-r)1 D(t) • The four gametes randomly unite to form a zygote. The proportion 1-r of the gametes produced by this zygote are parental (or nonrecombinant) gametes and fraction r are nonparental (or recombinant) gametes. A particular gamete, say AB, has a proportion (1-r) in generation t+1 produced without recombination. The frequency with which this gamete is produced in this way is (1-r)pAB(t). • Also this gamete is generated as a recombinant from the genotypes formed by the gametes containing allele A and the gametes containing allele B. The frequencies of the gametes containing alleles A or B are pA(t) and pB(t), respectively. So the frequency with which AB arises in this way is rpA(t)pB(t). • Therefore the frequency of AB in the generation t+1 is pAB(t+1) = (1-r)pAB(t) + rpA(t)pB(t) By subtracting is pA(t)pB(t) from both sides of the above equation, we have D(t+1) = (1-r)1 D(t) Whence D(t+1) = (1-r)t D(1) Estimate and test for LD Assuming random mating in the population, we have joint probabilities of the two genes BB (PBB) Bb (PBb) bb (Pbb) _______________________________________________________________________________________ AA (PAA) pAB2 2PABPAb PAb2 n22 n21 n20 Aa (PAa) 2PABPaB 2(PABPab+PAbPaB) 2PAbPab n12 n11 n10 2 aa (Paa) PaB 2PAbPab Pab2 n02 n01 n00 ________________________________________________________________________________________ Multinomial pdf H1: D 0 log f(pij|n) = log n!/(n22!…n00!) + n22 log pAB2 + n21log (2pABpAb) + n20 log pAb2 +… Estimate pAB, pAb, paB (pab = 1-pAB-pAb-paB) pA, pB, D H0: D = 0 log f(pi,pj|n) = log n!/(n22!…n00!) + n22log(pApB)2 + n21log(2pA2pBpb)+n20log(pApb)2 +… Estimate pA and pB. Chi-square Test of Linkage Disequilibrium (D) Test statistic x2 = 2nD2/(pApapBpb) is compared with the critical threshold value obtained from the chi-square table x2df=1 (0.05). n is the number of individuals in the population. If x2 < x2df=1 (0.05), this means that D is not significantly different from zero and that the population under study is in linkage equilibrium. If x2 > x2df=1 (0.05), this means that D is significantly different from zero and that the population under study is in linkage disequilibrium. Example (1) Two genes A with allele A and a, B with alleles B and b, whose population frequencies are denoted by pA, pa (=1- pA) and pB, pb (=1- pb), respectively (2) These two genes are associated with each other, having the coefficient of linkage disequilibrium D Four gametes are observed as follows: Gamete AB Obs 474 Gamete frequency pAB Ab 611 pAb aB 142 paB ab 773 pab Total 2n=2000 =474/2000 =611/2000 =142/2000 =773/2000 =0.237 =0.305 =0.071 =0.386 1 Estimates of allele frequencies pA = pAB + pAb = 0.237 + 0.305 = 0.542 pa = paB + pab = 0.071 + 0.386 = 0.458 pB = pAB + paB = 0.237 + 0.071 = 0.308 pb = pAb + pab = 0.305 + 0.386 = 0.692 The estimate of D D = pABpab – pAbpaB = 0.237 0.386 – 0.305 0.071 = 0.0699 Test statistics x2 = 2nD2/ (pApapBpb) =210000.06992/(0.5420.4580.3080.692) = 184.78 is greater than x2df=1 (0.05) = 3.841. Therefore, the population is in linkage disequilibrium at these two genes under consideration. A second approach for calculating x2: Gamete Obs Exp AB 474 2n(pApB) =334.2 Ab aB 611 142 2n(pApb) 2n(papB) =750.8 =281.8 ab 773 2n(papb) =633.2 Total 2n=2000 2000 x2 = (obs – exp)2 /exp = (474-334.2)2/334.2 + (611-750.8)2/750.8 + (142-281.8)2/281.8 + (773-633.2)2/633.2 = 184.78 = 2nD2/ (pApapBpb) Measures of linkage disequilibrium (1) D, which has a limitation that its value depends on the allele frequencies • • D = 0.02 is considered to be large for two genes each with diverse allele frequencies, e.g., pA = pB = 0.9 vs. pa = pb = 0.1 small for two genes each with similar allele frequencies, e.g., pA = pB = 0.5 vs. pa = pb = 0.5 (2) To make a comparison between gene pairs with different allele frequencies, we need a new normalized measure. The range of LD is max(-pApB, -papb) D min(pApb, papB) The normalized LD (Lewontin 1964) is defined as D' = D/ Dmax, where Dmax is the maximum that D can have, which is Dmax = max(-pApB, -papb) if D < 0, or min(pApb, papB) if D > 0. (3) Linkage disequilibrium measured as the correlation between the A and B alleles R = D/(pApapBpb), r: [-1, 1] Note: x2= 2nR2 follows the chi-square distribution with df = 1 under the null hypothesis of D = 0. For the above example, we have R = 0.0699/(pApbpapB) = 0.3040. Application of LD analysis D(t+1) = (1-r)tD(t), This means that when the population undergoes random mating, the LD decays exponentially in a proportion related to the recombination fraction. (1) Population structure and evolution Estimating D, D' and R the mating history of population The larger the D’ and R estimates, the more likely the population in nonrandom mating, the more likely the population to have a small size, the more likely the population to be affected by evolutionary forces. Human origin studies based on LD analysis Reich, D. E., M. Cargill, S. Bolk, J. Ireland, P. C. Sabeti, D. J. Richter, T. Lavery, R. Kouyoumjian, S. F. Farhadian, R. Ward and E. S. Lander, 2001 Linkage disequilibrium in the human genome. Nature 411: 199-204. Dawson, E., G. R. Abecasis, S. Bumpstead, Y. Chen et al. 2002 A first-generation linkage disequilibrium map of human chromosome 22. Nature 418: 544-548. LD curve for Swedish and Yoruban samples. To minimize ascertainment bias, data are only shown for marker comparisons involving the core SNP. Alleles are paired such that D' > 0 in the Utah population. D' > 0 in the other populations indicates the same direction of allelic association and D' < 0 indicates the opposite association. a, In Sweden, average D' is nearly identical to the average |D'| values up to 40-kb distances, and the overall curve has a similar shape to that of the Utah population (thin line in a and b). b, LD extends less far in the Yoruban sample, with most of the long-range LD coming from a single region, HCF2. Even at 5 kb, the average values of |D'| and D' diverge substantially. To make the comparisons between populations appropriate, the Utah LD curves are calculated solely on the basis of SNPs that had been successfully genotyped and met the minimum frequency criterion in both populations (Swedish and Yoruban) (Reich,te al. 2001) (2) Fine mapping of disease genes The detection of LD may imply that the recombination fraction between two genes is small and therefore closer (given the assumption that t is large). Quantitative genetics • • • • • • • • Many traits that are important in agriculture, biology and biomedicine are continuous in their phenotypes. For example, Crop Yield Stemwood Volume Plant Disease Resistances Body Weight in Animals Fat Content of Meat Time to First Flower IQ Blood Pressure The following image demonstrates the variation for flower diameter, number of flower parts and the color of the flower Gaillaridia pilchella (McClean 1997). Each trait is controlled by a number of genes each interacting with each other and an array of environmental factors. Number of Genes Number of Genotypes 1 2 5 10 3 9 243 59,049 Consider two genes, A with two alleles A and a, and B with two alleles B and b. - Each of the alleles will be assigned metric values - We give the A allele 4 units and the a allele 2 units - At the other locus, the B allele will be given 2 units and the b allele 1 unit Genotype AABB AABb AAbb AaBB AaBb Aabb aaBB aaBb aabb Ratio 1 2 1 2 4 2 1 2 1 Metric value 12 11 10 10 9 8 8 7 6 A grapical format is used to present the above results: Normal distribution of a quantitative trait may be due to • Many genes • Environmental effects The traditional view: polygenes each with small effect and being sensitive to environments The new view: A few major gene and many polygenes (oligogenic control), interacting with environments Traditional quantitative genetics research: Variance component partitioning • The phenotypic variance of a quantitative trait can be partitioned into genetic and environmental variance components. • To understand the inheritance of the trait, we need to estimate the relative contribution of these two components. • We define the proportion of the genetic variance to the total phenotypic variance as the heritability (H2). - If H2 = 1.0, then the trait is 100% controlled by genetics - If H2 = 0, then the trait is purely affected by environmental factors. • Fisher (1918) proposed a theory for partitioning genetic variance into additive, dominant and epistatic components; • Cockerham (1954) explained these genetic variance components in terms of experimental variances (from ANOVA), which makes it possible to estimate additive and dominant components (but not the epistatic component); • I proposed a clonal design to estimate additive, dominant and part-of-epistatic variance components Wu, R., 1996 Detecting epistatic genetic variance with a clonally replicated design: Models for low- vs. high-order nonallelic interaction. Theoretical and Applied Genetics 93: 102-109. Genetic Parameters: Means and (Co)variances One-gene model Genotype Genotypic value Net genotypic value aa G0 -a 0 Aa G1 d AA G2 a origin=(G0+G1)/2 a = additive genotypic value d = dominant genotypic value Environmental deviation E0 E1 E2 Phenotype or Phenotypic value Y0=G0+E0 Y1=G1+E1 Y2=G2+E2 Letting =a+(q-p)d P0 =q2 -a - =-2p[a+(q-p)d] -2p2d =-2p-2p2d P1 =2pq d- = (q-p)[a+(q-p)d] +2pqd =(q-p)+2pqd P2 =p2 a- = 2q[a+(q-p)d] -2q2d =2q-2q2d Breeding value Dominant deviation -2p -2p2d (q-p) 2pqd 2q -2q2d Genotype frequency at HWE Deviation from population mean Population mean = q2(-a) + 2pqd + p2a = (p-q)a+2pqd Genetic variance 2g = q2(-2p-2p2d)2 + 2pq[(q-p)+2pqd]2 + p2(2q-2q2d)2 = 2pq2 + (2pqd)2 = 2a (or VA) + 2d (or VD) Additive genetic variance, Dominant genetic variance, depending on both on a and d depending only on d Phenotypic variance 2P = q2Y02 + 2pqY12 + p2Y22 – (q2Y0 + 2pqY1 + p2Y2)2 Define H2 = 2g /2P as the broad-sense heritability h2 = 2a / 2P as the narrow-sense heritability These two heritabilities are important in understanding the relative contribution of genetic and environmental factors to the overall phenotypic variance. What is = a+(q-p)d? It is the average effect due to the substitution of gene from one allele (A say) to the other (a). Event A a contains two possibilities Frequency Value change From Aa to aa q d-(-a) a-d = q[d-(-a)]+p(a-d) = a+(q-p)d From AA to Aa p Midparent-offspring correlation ____________________________________________________________________ Progeny Genotype Freq. of Midparent AA Aa aa Mean value of parents matings value a d -a of progeny ____________________________________________________________________ AA × AA p4 a 1 a AA × Aa 4p3q ½(a+d) ½ ½ ½(a+d) AA × aa 2p2q2 0 1 d Aa × Aa 4p2q2 d ¼ ½ ¼ ½d Aa × aa 4pq3 ½(-a+d) ½ ½ ½(-a+d) aa × aa q4 -a 1 -a ________________________________________________ Covariance between midparent and offspring: Cov(OP¯) = E(OP¯) – E(O)E(P¯) = p4a a + 4p3q ½(a+d) ½(a+d) + … + q4 (-a)(-a) – [(p-q)a+2pqd]2 = pq2 = ½2a The regression of offspring on midparent values is b = Cov(OP¯)/2(P¯) = ½2a / ½2P = 2a /2P = h2 where 2(P¯)=½2P is the variance of midparent value. IMPORTANT The regression of offspring on midparent values can be used to measure the heritability! This is a fundamental contribution by R. A. Fisher. You can derive other relationships Degree of relationship Covariance ____________________________________________________ Offspring and one parent Cov(OP) = 2a/2 Half siblings Cov(FS) = 2a/4 Full siblings Cov(FS) = 2a/2 + 2a/4 Monozygotic twins Cov(MT) = 2a + 2d Nephew and uncle Cov(NU) = 2a/4 First cousins Cov(FC) = 2a /8 Double first cousins Cov(DFC) = 2a/4 + 2d/16 Offspring and midparent Cov(O) = 2a/2 ____________________________________________________