* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Zinc-Finger Proteins Required for Pairing and Synapsis
Saethre–Chotzen syndrome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Frameshift mutation wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene expression profiling wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Y chromosome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
X-inactivation wikipedia , lookup
Neocentromere wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
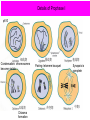
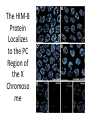
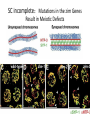
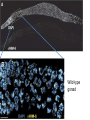
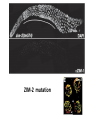
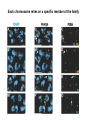
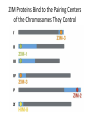
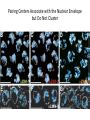
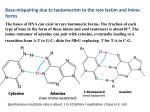
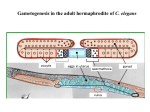
A Zinc-Finger Protein family Required for Pairing and Synapsis in C. elegans 杨茹&岳云欢 Review • Pairing: side-by-side alignment of homologs • Synapsis: intimate association of chromosomes specifically in the context of the synaptonemal complex (SC) Details of Prophase I p610 细线期 Condensation: chromosomes become visible 双线期 Chiasma formation 偶线期 Pairing: telomere bouquet 终变期 粗线期 Synapsis is complete 中期I Former investigation The wild-type haploid karyotype of C. elegans HIM-8 protein • HIM:high incidence of males • HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051–1063. [Phillips, C.M., Wong, C., Bhalla, N., Carlton, P.M., Weiser, P.,Meneely, P.M., and Dernburg, A.F. (2005).] The HIM-8 Protein Localizes to the PC Region of the X Chromoso me New Results • The Gene cluster of the Family of Zinc-Finger Proteins ZIM: zinc-finger protein in meiosis SC incomplete:Mutations in the zim Genes Result in Meiotic Defects Results of Mutation of Each zim Gene Each zim Gene Is Responsible for Synapsis of Specific Autosome Pairs Diagram of a hermaphrodite gonad 1.Premeiotic 2.Pairing and Synapsis 3. 4. &5.pachtene Pairing of Chromosome V Is Disrupted in the zim-2 Mutant Diagram of a hermaphrodite gonad Genomic locations of FISH Wild-type gonad premeiotic Transition zone Pachytene region Wild-type gonad ZIM-2 mutation Each chromosome relies on a specific member of the family ZIM Proteins Bind to the Pairing Centers of the Chromosomes They Control Pairing Centers Associate with the Nuclear Envelope but Do Not Cluster Multiple foci dispersed along the nuclear periphery •Schematic of the ZIM/HIM-8 gene cluster in C. elegans, C. briggsae, and C. remanei. Predicted orthologs based on sequence comparisons are linked by gray shading. Evolution of the ZIM/HIM-8 Protein Family Questions • The relationship between zim/him and telomeres? • Recognition of the homolog chromosome Conclusion & Review Is the result important? The first effort to find a family of protein working in pairing and synapsis Why could they succeed? The good model animal: C.elegans The former research : SC、PC、HIM-8 ……