* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Synthetic lethal analysis of Caenorhabditis elegans posterior
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Behavioural genetics wikipedia , lookup
Oncogenomics wikipedia , lookup
Population genetics wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic testing wikipedia , lookup
RNA silencing wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genomic imprinting wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Synthetic biology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Public health genomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Essential gene wikipedia , lookup
Pathogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Microevolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
History of genetic engineering wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome (book) wikipedia , lookup
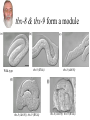
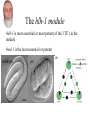
Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions L Ryan Baugh, Joanne C Wen, Andrew A Hill, Donna K Slonim, Eugene L Brown & Craig P Hunter* The Hunter Lab at Harvard Dr. Craig Hunter •Examining the mechanism behind systemic RNAi •Studying the master switch pal-1, involved in specifying the fate of the C blastomere Background •Most genes are not essential (i.e. yeast, flies, worms) •2 possible reasons: homology (direct compensation) & parallel pathways (indirect compensation) •Genes with 1 or more homologs less likely to have loss-offunction phenotype •2/3 genetic buffering due to homology, implies large role for parallel pathways How do you characterize mechanisms of phenotypic robustness? Background: Synthetic Lethality •Developed by Charlie Boone at U of T •SGA= systematic construction of double mutants •Cross YFG1 to an array of ~ 5000 Δ strains Synthetic Lethality •Identify functional relationships between genes & pathways •Shed light on how regulatory networks buffer gene function •Allows for creation of genetic modules •Helps identify nodes The C Blastomere pal-1 specifies & regulates C lineage •PAL-1 target genes RNAi of most PAL-1 targets does not result in•Identified a in phenotype. Why? Is there overlapping function? microarray screen •Validated using GFP transcriptional reporters •Many targets are TF’s Goal of paper •Identify synthetic interactions between pairs of PAL-1 targets •Determine if genetic modules exist that buffer loss of proteins in the pal-1 pathway •Examine the feasibility/reproducibility of double RNAi experiments Experimental Methods RNAi: Soaked strains of worms in dsRNA (100-1000bp) -Added minimal T7 promoter to PCR primers -Amplify DNA for in vitro transcription -dsRNA reannealed by heating & cooling Attempted assembling matrix with only RNAi -led to variable, inconsistent results** Examined RNAi-treated progeny for % embryonic lethality -converted % lethality to % survival to calculate significance of the interaction Statistical Analysis 1. % lethality converted to % survival 2. Survival values normalized 3. Calculate significance of interactions using students t test (p>0.05) Ho: Survival of the double disruption (mutation x RNAi) equals the product of survivals for each single disruption Results Which interactions are significant? interaction interaction tbx-8 & tbx-9 form a module Wild-type tbx-9 (RNAi) tbx-8 (ok656); tbx-9 (RNAi) tbx-8 (ok656) tbx-8 (ok656); tbx-9 (RNAi) tbx-8 & tbx-9 form a module •Either disruption on their own: 1-5% lethality, 5% of hatchlings display posterior body defects •Double disruption results in 50% lethality; severe defects in hatchlings •tbx-8,9 interactions are conserved in C. briggsae & display similar expression patterns; thus module has likely been conserved A muscle differentiation module •Identified a module around hlh-1 •Detected 5 of 6 interactions (p<0.001) between hnd-1, hlh-1 and unc-120 •Disruption of any combination of the 3 genes results in pat •Some symmetry, but not interactions are symmetrical. Why? The hlh-1 module •hlh-1 is most essential (or most potent) of the 3 TF’s in the module •hnd-1 is the least essential (or potent) wildtype pat Is this module conserved? •Interactions between bHLH factors in vertebrates •Relationship between bHLH proteins & MADS-box regulators (the MEF2 group) Criticisms •No positive controls (i.e. no known interactions were used) •Why choose soaking and not do a comparison to feeding & injecting? •Why limit the interactions to lethality? Why not search for enhancement of phenotypes (gro, lva, lvl etc.) •Didn’t confirm results by doing a dihybrid cross Gratuitous Political Cartoons The People have spoken! Plebiscite results 1 0.9 0.8 0.7 percentile 0.6 Property Food/beer? 0.5 0.4 0.3 0.2 0.1 0 1 2