* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download ppt
Group selection wikipedia , lookup
Koinophilia wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Heritability of autism wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genetic testing wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Genetic drift wikipedia , lookup
Designer baby wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
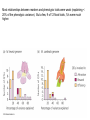
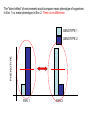
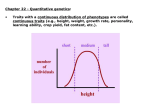
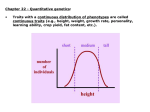
IV. Variation in Quantitative Traits A. Quantitative Effects IV. Variation in Quantitative Traits A. Quantitative Effects - the more factors that influence a trait (genetic and environmental) , the more 'continuously variable' the variation in that trait will be. IV. Variation in Quantitative Traits A. Quantitative Effects - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). A two gene trait with additive effects (height ‘dose’) can make 5 phenotypes (‘dose’ = 0, 1, 2, 3, 4), and so forth. IV. Variation in Quantitative Traits A. Quantitative Effects - the more genes that influence a trait, the more 'continuously variable' the variation in that trait will be. - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). AA, Aa, aa (Tall, medium, short) However, a two-gene trait with incomplete dominance at both loci can have nine variants: AA, Aa, aa X BB, Bb, bb - So, as the number of genes affecting a trait increase, the variation possible can increase multiplicatively. IV. Variation in Quantitative Traits A. Quantitative Effects - the more genes that influence a trait, the more 'continuously variable' the variation in that trait will be. - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). AA, Aa, aa (Tall, medium, short) However, a two-gene trait with incomplete dominance at both loci can have nine variants: AA, Aa, aa X BB, Bb, bb -So, as the number of genes affecting a trait increase, the variation possible can increase multiplicatively. -If there are environmental effects, then the distribution of phenotypes can be continuous. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits 1. Quantitative Trait Loci (QTL) mapping Monkeyflowers – genus Mimulus (Bradshaw et al. 1998) Occur in the Sierras, with overlapping elevational distributions. They readily hybridize, but no hybrids are found in nature – possibly because they attract different pollinators. The species vary in flower shape and structure. Since they each “breed true” for their particular phenotypes, we assume they are homozygous at loci influencing these traits. Since they hybridize, we can form heterozygous F1’s (b). Mating F1’s creates F2’s, which show quantitative variation in color, shape, and nectar volume (12 measured traits, like corolla width, stamen length, anthocyanins in petal, etc.) M. Lewisii F1 M. cardinalis Identify ‘marker’ loci across the genome – loci that are: 1 - unique in the genome 2 – homozygous in each species 3 – distributed over all chromosomes M. Lewisii F1 M. cardinalis Identify ‘marker’ loci across the genome – loci that are: 1 - unique in the genome 2 – homozygous in each species 3 – distributed over all chromosomes The Goal: Find correlations between F2 ‘marker’ genotypes and either color or shape. Many genes with small effects or a few genes with large effects? So, maybe (j) and (g) are homozygous with the M. lewisii allele at markers 1and 5, while (l) is homozygous for the M. cardinalis marker allele at markers 1 and 5. This correlation between marker genotype and parental phenotype suggests that there are QTL’s for color near markers 1 and 5. M. Lewisii F1 M. cardinalis Identify ‘marker’ loci across the genome – loci that are: 1 - unique in the genome 2 – homozygous in each species 3 – distributed over all chromosomes The Goal: So, we have used the concept of linkage (and “linkage disequilibrium”, really) between a marker locus and phenotypic trait to isolate a region that influences the trait. In addition, the strength of the relationship (the amount of phenotypic variance explained by differences in genotypes), describes the strength of the gene’s effect. M. Lewisii F1 M. cardinalis Most relationships between markers and phenotypic traits were weak (explaining < 20% of the phenotypic variance). But a few, 9 of 12 floral traits, %’s were much higher. One marker explained over 80% of the variation in color. Genotypes varying ONLY at this locus and are visited by bees and hummingbirds, respectively. Single genes can exert strong effects that can be driven quickly to fixation by selection. Determining the actual gene in the region, and the protein and action of the protein influencing the trait, requires additional “genetic dissection” of ‘Candidate Loci’ in the region. IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen) IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen) - Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g) - Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g) GENOTYPE 1 AND THESE CAN BE VERY IMPORTANT: PHENOTYPE GENOTYPE 2 ENV 1 ENV 2 The "direct effect" of environment would compare mean phenotype of organisms in Env 1 vs. mean phenotype in Env 2. There is no difference. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2 The "direct effect" of 'genotype' would compare mean phenotype of Genotype 1 vs. mean phenotype of Genotype 2. There is no difference. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2 But there is a SIGNIFICANT "genotype x environment" interaction. The effect of environment on the phenotype depends on the genotype. This important component of variation is often obscured in simplistic direct models. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2 IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen) - Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g) - Ultimately, the goal of evolutionary studies is to determine the contribution of genetic variation, because this is the only variation that is heritable and can evolve. “Broad-sense” heritability = Vg/Vp IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation - Even the genetic variation is more complex than one might think. There is variation due to 'additive' genetic variance, 'dominance' genetic variance, 'epistasis', and a variety of other contributors (sex linkage) that can be modeled. V(g) = V(a) + V(d) + V(ep) IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation - Even the genetic variation is more complex than one might think. There is variation due to 'additive' genetic variance, 'dominance' genetic variance, 'epistasis', and a variety of other contributors that can be modeled. - We will concern ourselves with 'additive variation' Think of an individual that is AA. If the 'A' allele is adaptive, then their fitness will be higher than the mean fitness of the population. Their offspring, as a consequence of inheriting this adaptive gene, will also have a higher fitness than the population, as a whole. This allele 'adds' fitness. 2 A’s (AA) adds more… IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure - Narrow-sense (h2) = V(a)/V(p) IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure - Narrow-sense (h2) = V(a)/V(p) – easier to measure Calculate the average phenotype of two parents, and calculate the average phenotype of their offspring. Graph these points across sets of parents and their offspring. The slope of the best-fit line (least-squares linear regression) describes the strength of the “heritability” of the trait. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0.8. because R = h2 s IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0.8. - The closer the offspring are to their particular parents (in amount of deviation from the whole population), the greater the heritability and the more rapid the response to selection. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0.8. - The closer the offspring get to their particular parents (in amount of deviation from the whole population), the greater the heritability and the more rapid the response to selection. - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember? IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure. As selection proceeds, most variation is environmental or dominance and response to selection slows. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure. As selection proceeds, most variation is environmental or dominance and response to selection slows. - So, counterintuitively, adaptive traits may show low heritability...they have already been selected for, and most of the phenotypic variation NOW is probably environmental. EXAMPLE: Polar bears all have genetically determined white fur - it has been adaptive and has become fixed in their population. But they still vary in coat color (phenotype) as a result of dirt, etc. But the offspring of dirty bears will be just as white as the offspring of clean bears... no response to selection for 'dirty bears' because all the variation is environmental at this point. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments 4. Misuses of Heritability: Heritability is a property of a trait, in a given population, in a given environment. It provides no insight for comparisons across populations in different environments…. Why? IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability is a property of a trait, in a given population, in a given environment. It provides no insight for comparisons across populations in different environments…. Why? Genotype x environment interactions… Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “0” (same environment). Can we use these data to predict how these genotypes would grow, relative to one another, in another environment? Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “0” (same environment). No. Although there is high heritability in BOTH populations for plant height. Can we use these data to predict how these genotypes would grow, relative to one another, in another environment? Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “0” (same environment). IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability DOES NOT equal “genetically based” Many traits that are determined genetically are fixed, with no genetic variation, and so have very low heritability. IV. Variation in Quantitative Traits A. Quantitative Effects B. Identifying Loci Contributing to Quantitative Traits C. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability DOES NOT equal “genetically based” 1) Many traits that are determined genetically are fixed, with no genetic variation, and so have very low heritability. 2) Heritability is measured on one population in one environment. You cannot ascribe phenotypic variation BETWEEN groups – especially if they are in different environments, to ‘heritability’ and genetic factors. Ethnic groups differ in mean I.Q. And when measured in one population, I.Q. is heritable. But that doesn’t mean that “genetic differnces” explain the differrence between ethic groups in this trait…. Especially because environments vary. MZ-DZ twin studies: Vp = Vg + Ve - MZ twins: Vg = 0, so Vp for a trait = only Ve. twin studies: Some social psychologists believe that we can determine “heritability” or “genetic contribution” (!) to a trait by examining the degree of similarity between ‘monozygotic’ (identical) and ‘dizygotic’ (fraternal) twins. MZ-DZ twin studies: Vp = Vg + Ve - MZ twins: Vg = 0, so Vp for a trait = only Ve. - DZ twins: Us DZ twins to measure Vg = Vp – Ve (mz) - problem: MZ twins are often treated more alike than DZ twins. So, many of their similarities may be environmental, too. Thus, Ve is underestimated. - when this artificially LOW Ve is subtracted from Vp for DZ twins, it OVERESTIMATES the genetic contribution to that trait. For MZ twins, clothes choice shows very little variation. (Ve = 0.1). MZ-DZ twin studies: Vp = Vg + Ve - MZ twins: Vg = 0, so Vp for a trait = only Ve. - DZ twins: Us DZ twins to measure Vg = Vp – Ve (mz) - problem: MZ twins are often treated more alike than DZ twins. So, many of their similarities may be environmental, too. Thus, Ve is underestimated. - when this artificially LOW Ve is subtracted from Vp for DZ twins, it OVERESTIMATES the genetic contribution to that trait. For MZ twins, clothes choice shows very little variation. (Ve = 0.1). DZ twins dress different (Vp = 10.0). Vg = Vp – Ve = 10.0 – 0.1 = 9.9 H2 for ‘clothes wearing’ = Vg/Vp = 9.9/10.0 = 0.99. WOW! WHAT A HUGE GENETIC CONTRIBUTION!!! MZ-DZ twin studies: Vp = Vg + Ve Hmmmm… MZ twins are treated more similarly than DZ twins in their homes, so Ve differs between the groups. Hmmmm…. Suppose we compare MZ and DZ twins reared apart, through adoption? Then Ve will be the same across groups, and greater similarity among MZ twins must be a function of greater genetic similarity. MZ DZ “The Jim Twins” Ve is the same for both groups • As youngsters, each Jim had a dog named "Toy." • Each Jim had been married two times -- the first wives were both called "Linda" and the second wives were both called "Betty." • One Jim had named his son "James Allan" and the other Jim had named his son "James Alan." • Each twin had driven his light-blue Chevrolet to Pas Grille beach in Florida for family vacations. • Both Jims smoked Salem cigarettes and drank Miller Lite beer. • Both Jims had at one time held part-time posts as sheriffs. • Both were fingernail biters and suffered from migraine headaches. • Each Jim enjoyed leaving love notes to his wife throughout the house. I.Q.: Statistically significant differences in mean performances of ethnic groups in U.S. Also, I.Q. (as measured in single populations) is heritable. “Most scholars accept that I.Q. in the human species as a whole is substantially heritable, somewhere between 40 percent and 80 percent, meaning that much of the observed variation in I. Q. is genetic.” – Murray and Herrnstein (1994). No. And even if so, so what? Much of the variation is environmental.