* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Nucleotides and Nuclic Acids
Genetic engineering wikipedia , lookup
DNA barcoding wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
DNA sequencing wikipedia , lookup
Holliday junction wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Restriction enzyme wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA vaccination wikipedia , lookup
Transformation (genetics) wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Molecular cloning wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
History of genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
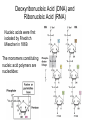
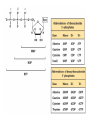
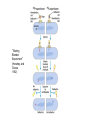
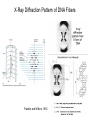
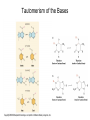
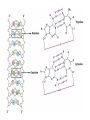
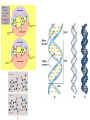
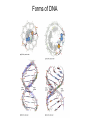
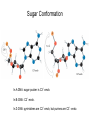
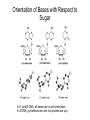
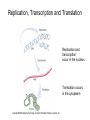
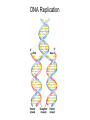
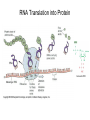
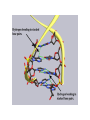
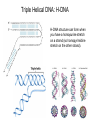
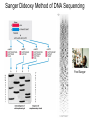
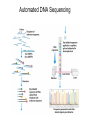
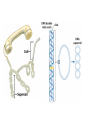
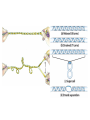
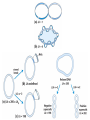
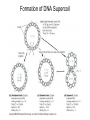
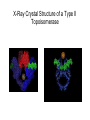
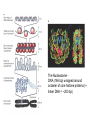
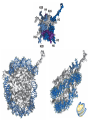
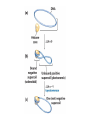
Nucleotides and Nucleic Acids The Basics Deoxyribonucleic Acid (DNA) and Ribonucleic Acid (RNA) Nucleic acids were first isolated by Friedrich Miescher in 1869. The monomers constituting nucleic acid polymers are nucleotides: Purine and Pyrimidine Bases in DNA and RNA Deoxyribonucleotides Ribonucleotides The Bases Absorb UV Light DNA is the Genetic Material “Transforming principle” of pneumonia-causing bacteria (Griffith, 1928) DNA is the “transforming principle” (Avery, McLeod and McCarty, 1944) “Waring Blender Experiment” (Hershey and Chase, 1952) Avery, McLeod and McCarty, 1944 “Waring Blender Experiment” (Hershey and Chase, 1952) Three-Dimensional Structure of Deoxyribonucleic Acid (DNA) Double Helix: 50 Years of DNA http://www.nature.com/nature/dna50/ An Early Conjecture about DNA Structure Tetranucleotide hypothesis (Levene, 1910) X-Ray Diffraction Pattern of DNA Fibers Franklin and Wilkins, 1953 Rosalind Franklin Maurice Wilkins X-Ray Diffraction Pattern of DNA Fibers Franklin and Wilkins, 1953 Incorrect Triple-Helical Structure Pauling and Corey, 1953 Chargaff’s Rules: A=T and G=C purines = pyrimidines Chargaff’s Rules Tautomerism of the Bases April 25, 1953 MOLECULAR STRUCTURE OF NUCLEIC ACIDS A Structure for Deoxyribose Nucleic Acid We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel features which are of considerable biological interest. … It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material. … J. D. WATSON F. H. C. CRICK Medical Research Council Unit for the Study of Molecular Structure of Biological Systems, Cavendish Laboratory, Cambridge. Watson-Crick Base Pairs Elements of Structure in DNA Double Helix The DNA Double Helix Forms of DNA The A, B and Z Forms of DNA •Most DNA in the cell is in the B form. •A small amount of the DNA in the cell may locally adopt the Z conformation (where sequence has alternating purine and pyrimidine bases). •The A form of DNA is found in dehydrated samples of DNA but not normally in the cell. However, double-stranded RNA and DNARNA hybrids form helices resembling the A form of DNA. Z-DNA: Left-Handed Helix Alexander Rich, 1979 Proteins that Bind Z-DNA X-Ray structure of two ADAR1 Z domains in complex with Z-DNA. Nucleotide Conformation Sugar-phosphate backbone is conformationally constrained (O4’ generally gauche to O5’ since torsion angle about the C4’-C5’ bond is restricted). Sugar Conformation In A-DNA: sugar pucker is C3’-endo. In B-DNA: C2’-endo. In Z-DNA: pyrimidines are C2’-endo, but purines are C3’-endo. Orientation of Bases with Respect to Sugar In A- and B-DNA, all bases are in anti orientation. In Z-DNA, pyrimidines are anti, but purines are syn. Local Deviations from the Idealized B-DNA Structure Properties of Naturally Some Occurring DNA Molecules Haploid human genome: 23 chromosomes of ~5 × 107 bp to ~25 × 107 bp each for a total of ~3 × 109 bp. Chargaff’s Rules Redux DNA Replication and Transcription into RNA Genetic Information Flow: DNA RNA Protein “The Central Dogma” Francis Crick, 1957 Replication, Transcription and Translation Replication and transcription occur in the nucleus Translation occurs in the cytoplasm Polynucleotide Synthesis DNA Replication DNA Replication Hydrolysis of Pyrophosphate DGo’ = -33.5 kJ/mol Three Possible Models of DNA Replication Meselson-Stahl Experiment: Semiconservative DNA Replication Gene Expression (Transcription, Splicing and Translation) Expression of a hypothetical eukaryotic gene DNA Transcription into RNA Regulation of Gene Expression Catabolite activator protein (CAP)-cAMP, a bacterial transcription factor, bound to DNA Example of DNA Sequence Recognition by a DNA-Binding Protein 434 phage repressor protein mRNA Splicing RNA Translation into Protein The Genetic Code Ribosomal Peptidyl Transferase Activity Note: the catalytic component of the ribosome’s peptidyl transferase activity is RNA; it’s an example of a catalytic RNA or ribozyme. Stability of Nucleic Acids The DNA Duplex Can Be Reversibly Denatured (Melted) Factors Determining DNA Duplex Stability In order for the double helix to form under given conditions, forces favoring duplex formation must outweigh disfavoring factors, so that the ∆G for duplex formation under those conditions is negative. Factors Stabilizing the DNA Duplex 1. “Hydrophobic interactions,” base stacking (vertical base stacking interactions make duplex formation enthalpically favored, although entropically opposed, unlike the Hydrophobic Effect involved in protein folding and lipid bilayer formation) 2. Ionic interactions (duplex becomes more stable as ionic strength increases, since presence of positive counterions partially neutralizes negative charges of backbone phosphates) 3. Hydrogen bonding between base pairs Under conditions in which the duplex forms spontaneously, stabilizing forces outweigh destabilizing forces, so that ∆G of duplex formation is negative. London Dispersion Forces in Base Stacking (Pi-Pi Stacking) Factors Destabilizing the DNA Duplex 1. Higher entropy of denatured random coil (denaturation is temperature-dependent since relevant term is -T∆S in the Gibb’s free energy equation, ∆G = ∆H - T∆S) 2. Charge-charge repulsion of backbone phosphates (so absence of positive counterion would favor denaturation) Under denaturing conditions, destabilizing factors dominate over stabilizing, so that ∆G of denaturation becomes negative (and ∆G of duplex formation now has a positive sign). Denaturation of DNA Duplex Increasing T makes denaturation favorable Denaturation and renaturation of DNA duplex are cooperative processes. Hyperchromic Shift Stability of Double Helix and Melting Temperature Increase as GC Content Increases Melting Temperature (Tm): ∆G (at Tm) = ∆H - Tm∆S = 0 So: Tm = ∆H/∆S GC-rich duplex is enthalpically more stable than AT-rich: While ∆S of melting per bp is about the same for all DNA molecules, ∆H (melting GC-rich DNA) > ∆H (melting AT-rich DNA) Instability Due to Reactivity: Individual RNA Strands Chemically Less Stable than DNA 2’-Hydroxyl participates in basecatalyzed intramolecular nucleophilic attack on phosphorus, breaking the phosphodiester backbone and forming a 2’,3’ cyclic phosphate, with further hydrolysis to 2’- and 3’monophosphate products. Thymine in Place of Uracil Makes DNA More Genetically Stable Spontaneous deamination = mutagenic base substitution Base pairs with guanine A DNA repair enzyme called uracil-DNA glycosylase removes uracil in DNA and replaces it with cytosine, undoing the mutagenic damage. (Normal thymine in DNA is not removed.) Base pairs with adenine Other Nucleic Acid Structures Non-Watson-Crick Base Pairing, e.g., Hoogsteen Base Pairing Triple Helical DNA: H-DNA H-DNA structure can form when you have a homopurine stretch on a strand (so homopyrimidine stretch on the other strand). Self-Complementary Nucleic Acid Strands and Hairpins Transfer RNA (tRNA) Structure Palindromic DNA Sequences: Potential to Form Cruciform Structures (Double Hairpins) Some Palindromes: Madam, I’m Adam And E.T. saw waste DNA. No, Mel Gibson is a casino's big lemon. A man, a plan, a canal: Panama! Palindromes and Restriction Endonucleases Another reason palindromes are important: Type II restriction enzymes are site-specific endonucleases used in molecular biology research (such as gene cloning) that recognize specific palindromic DNA sequences. X-ray crystal structure of Eco RI bound to DNA DNA cleavage products: Sticky ends (e.g., Eco RI): 5’-G-3’ 5’-AATTC-3’ 3’-CTTAA-5’ 3’-G-5’ Blunt ends (e.g., Sma I): 5’-CCC-3’ 5’-GGG-3’ 3’-GGG-5’ 3’-CCC-5’ Sanger Dideoxy Method of DNA Sequencing The most commonly used of two methods for DNA sequencing. The other method, not as used today, is the Maxam-Gilbert chemical cleavage method. NTP, dNTP and ddNTP (Dideoxynucleoside Triphosphate) Structures Sanger Dideoxy Method of DNA Sequencing Fred Sanger Automated DNA Sequencing DNA Topology and Supercoiling Relaxed vs. Supercoiled DNA Positive and Negative Supercoiling positive supercoil = left-handed = overwound DNA negative supercoil = right-handed = underwound DNA L=T+W • L or Lk = linking number (number of times one strand crosses the other) • T = twist (number of helical turns; for B-DNA, T = # bp divided by ~10.5 bp/turn) • W = writhe (number of supercoils) Superhelical Density: s = DL/L0 = W/T (L0 = linking number of relaxed molecule = T, since W = 0 in relaxed molecule) Formation of DNA Supercoil Supercoiling of DNA Shown by Gel Electrophoresis Agorose Gel Stained with Ethidium Bromide Supercoiled circular DNA migrates faster during agarose gel electrophoresis than does relaxed circular DNA (or linear DNA). Ethidium = fluorescent intercalating agent used to stain DNA in agarose gels Type I Topoisomerases Ο Ο Ο Ο •∆L = ±1 per cycle •Cleaves a single strand •Passes broken single strand around the other, then rejoins strands •Does not require ATP •Relaxes supercoiled DNA Structure of a Type I Topoisomerase Type II Topoisomerases •∆L = ±2 per cycle •Cleaves both strands •Passes unbroken part of duplex through doublestrand break, then rejoins strands •Requires ATP •Relaxes supercoiled DNA •Some type II enzymes (like DNA gyrase) can add negative supercoils Topological Interconversions Catalyzed by Type II Topoisomerase Relaxation Catenation and Decatenation Knotting and Unknotting X-Ray Crystal Structure of a Type II Topoisomerase Chromosomes and Chromatin: HigherOrder DNA Structure The Nucleosome DNA (146 bp) wrapped around octamer of core histone proteins (+ linker DNA = ~200 bp) Negative Supercoiling of DNA is Important Biologically • Negatively supercoiled DNA is equivalent to underwound DNA. This makes it easier to separate strands during replication and transcription. • In eukaryotes, formation of nucleosomes results in torsional strain in the DNA molecule (equivalent to ~1.5-1.8 supercoils/nucleosome particle theoretically; actual value is ~1), which is relieved by topoisomerases. This results in DNA that is negatively supercoiled once histone proteins are removed. • In prokaryotes, an enzyme called DNA gyrase (a type II topoisomerase) generates negative supercoils.