* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Shared character
Survey
Document related concepts
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genealogical DNA test wikipedia , lookup
History of RNA biology wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Genetic code wikipedia , lookup
Metagenomics wikipedia , lookup
DNA barcoding wikipedia , lookup
History of genetic engineering wikipedia , lookup
Koinophilia wikipedia , lookup
Transitional fossil wikipedia , lookup
Maximum parsimony (phylogenetics) wikipedia , lookup
Transcript
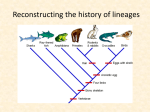
17-2 Systematics - Goal is to categorize organism by NATURAL relationships PHYLOGENETICS So systematic taxonomists think organism’s classficiation should reflect PHYLOGENY Phylogeny – evolutionary history of a species/taxon - Today’s taxonomists involved in phylogenetics – analyzing evolutionary/ancestral correlation between taxa Evidence to predict about phylogenetics o o o They compare visible similarities – between living and fossilized creatures Patterns in embryonic development - how diff species embryos can express similar genes Compare chromosomes, DNA, RNA, macromolecules from diff species This can be modeled through phylogenetic diagram/tree – shows how closely related some taxa can be.. keep in mind its subject to change when scientists find out new things! EVIDENCE OF SHARED ANCESTRY - Fossils are good for finding evolutionary changes, but there’s not much evidence for small/softbodied (worms, bacteria, fungi) o Can help with phylogenetic diagram, but systematist tests inferred relationships for more evidence Systematists will look at Homologous features ( sharing common ancestry) Jaws of pangolin and dog Analogous features (similar because of similar function, not lineage) Scales – pangolin& dog, but they both evolved independently in 2 taxa Embryological evidence Fluid-filled sac, amnion, surrounds embryos of birds/reptiles/mammals – this shared feature combines them into one taxon, excluding other vertebrates CLADISTICS - SYSTEM for phylogenetic analysis – using only shared/derived characterists to group taxa o o Shared character – one that all members of a group have in common – Derived character – one that evolved ONLY within group - Birds- feathers- only animals with feathers between living & fossils So the feathers were evolved only within bird line, not common ancestor Clade- group of organisms including ancestor and all descendants o No category names (class, phylum) Cladists make cladograms – their own phylogenetic diagrams o They have strict guidelines, so their taxonomies can be a little different from traditional Traditional taxonomists will place crocs with lizards and turtles in reptilia class, and birds in aves class But cladists put crocs and birds together in a clade because their common ancestor is more recent - Then it’s linked with successively later clades CONSTRUCTING A CLADOGRAM - Start with the outgroup – the organism distantly related to the other organisms 1. choose organisms—they go in left column, and put the outgroup there too. Top, going right, characteristics they could share 2. score the organisms lacking certain characteristic 0, and if it has it, score 1 3. most commonly shared derived character (this case vascular tissues) goes at the base of 1st branch of cladogram - 2nd most common; seeds, 3rd common; flowers - ferns lack seeds, that’s why they go on second branch Pine trees lack flowers; so third branch. And flowering plants go on last branch *so this tree is a cladistic hypothesis of the evolultionary relationship between these plants Cladists can also look at molecular level characteristics, or even a nucleotide in a gene sequence, or a protein’s amino acid sequence MOLECULAR CLADISTICS Take the common characteristics.. separate into thea diff charactersitsica – derived.. between species by making a tree Just by looking at the amino acid sequences, biologist can find relationships characteristics that makes it differ Molecular cladogram- branch lengths proportional to #of amino acid changes Shared characterists – the ones that everyone shares… - DOESN’T depend on any physical sims/diffs!! Molecular clock used to estimate sequence of past evolutionary events - Biologists use Evol. Changes of macromolecules (like DNA, RNA, proteins) as form of mol.. clock Molecular clock hypothesis - suggests that greater diff in sequences- more distant common ancestor - Carefully matched molecular clock with fossil records- can be used to hypothesize when characteristics arose and organisms diverged from their ancestors. CHROMOSOMES - Karyotypic data – doesn’t depend on molecular nor physical When the chromosomes are stained, showing pattern of bands, and those band have same pattern in same region of similar chromosomes, the regions are likely to have been inherited from 1 chrom. In last common ancestor of 2 species o When chromosomes have similar banding patterns, they may be homologous Here B2 and B3 together are supposed to seem homologous to A2 In this case, biologists can still hypothesize that they all came from common ancestor Pangolin classification – pangolins share adaptations with aardvarks&anteaters but they’re analogous. More analysis Scientists use all diff info to make their phylogenetic models shows their skeletal structure and nucleotide sequence are all on diff branches on this mammalian phylogenetic tree. So - Physical features the pangolins are put closer to dogs & bears than aard &anteaters - Embryos - Genes in nucleus - Mitochondrial DNA - Ribosomal RNA