* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 0 1 0 1 1 1 0 0 1 0
Human genome wikipedia , lookup
Genome (book) wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Metagenomics wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Behavioural genetics wikipedia , lookup
Population genetics wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Genetic drift wikipedia , lookup
Microevolution wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Human genetic variation wikipedia , lookup
Public health genomics wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
HLA A1-B8-DR3-DQ2 wikipedia , lookup
Haplogroup G-M201 wikipedia , lookup
A30-Cw5-B18-DR3-DQ2 (HLA Haplotype) wikipedia , lookup
Computational problems involving
Single Nucleotide Polymorphisms
Pritam Chanda
1
Agenda
•
•
•
•
•
•
Biological background
SNP representation
Tag SNP selection
Haplotype analysis
SNP-disease association study
Discussion
2
Central Dogma
3
A cell and its chromosomes
4
DNA structure
Base pairs : A-T, G-C
5’
A T A T A TG CA GC A
3’
3’
Template strand
5’
T A T A T AC GT CG T
Anti-parallel chain
Thus, each chromosome can be thought of as a sequence of A, T, G, C’s
5
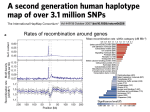
Genomic Variation and SNPs
• Human Genome 3 109 base pairs.
• Main form of variation between individual genomes: single
nucleotide polymorphisms (SNPs)
– Single base changes in the genome sequence that occurs in a
significant proportion (more than 1 percent) of the population
– Most SNPs are bi-allelic (two variations)
Sequences on a pair of
homologous chromosomes
• Total #SNPs 1 107
• Difference between any two individuals 3 106 SNPs ( 0.1% of
entire genome)
6
Why important ?
• A SNP (pronounced as ‘snip’) can alter the amino acid
sequence of the protein produced.
• Not always
– A protein consists of sequence of amino acids.
– There are total 20 amino acids
– Genetic code produces amino acids by reading groups of 3
nucleotides at a time
• 43 combinations = 64 different combinations of A,T,G,C.
– Thus not all combinations of 3 nucleotides produce different
amino acids
• Redundancy in genetic code.
– A SNP in which both alleles lead to the same protein sequence
is termed synonymous
– If different proteins are produced they are non-synonymous.
7
Why important ?
• SNPs that are not in protein coding regions may still
have consequences for gene splicing, transcription factor
binding, or the sequence of non-coding RNA.
• SNPs in humans can affect how humans develop
diseases, respond to pathogens, chemicals, drugs, etc.
• SNPs are inherited and do not change much from
generation to generation in an individual with time,
– SNPs are of great value to biomedical research and in
developing diagnostic and pharmaceutical products.
8
Bioinformatics representation
Sequences on a pair of homologous chromosomes
• Assumption: a snp is bi-allelic.
• Major allele
– most frequent allele
Sample 1
A G A T A G T A AT
A G A T C G T A AT
Sample 2
A G A T A G T A AT
A G A T A G T A AT
• Minor allele
– The other one
Sample 3
• Example
– Given DNA sequence
• Major allele (A) - 67%
• Minor allele (C) - 33%
Sample 1
A G A T 0 G T A AT
A G A T 1 G T A AT
Sample 2
A G A T 0 G T A AT
A G A T 0 G T A AT
• Encoding
– Major allele : 0
– Minor allele : 1
A G A T A G T A AT
A G A T C G T A AT
Sample 3
A G A T 0 G T A AT
A G A T 1 G T A AT
9
Haplotypes and Genotypes
•
Diploid organisms: cells have two homologous set of chromosomes.
•
Haplotype: description of SNP alleles on a single chromosome
– 0/1 vector, e.g., 00110101 (here, 0 is for major, 1 is for minor allele).
•
Genotype: combined description of SNP alleles on pairs of homologous
chromosomes
– 0/1/2 vector, e.g., 01122110 (0=0+0, 1=1+1, 2=0+1 or 1+0)
– Each genotype with k 2’s (heterozygotes) can be explained by 2k-1 pairs
of haplotypes
snps
Other nucleotides
A G A T A G T A AT
A C A T G G T A AA
Major allele
Minor allele
Haplotype
0 1 0 1 1
1 0 0 1 0
Heterozygous
Genotype
2 2 0 1 2
Homozygous
10
SNP databases
• HapMap project (www.hapmap.org)
– The aim of the project is to record the significant SNPs.
– Started in October 2002.
– Phase 1 data have been published and analysis of Phase 2 data is
underway as of October 2006.
• dbSNP
– A database of SNPs and short deletion and insertion polymorphisms at
NCBI.
• CGAP
– Genetic variation in genes important in cancer (At the National Cancer
Institute)
• EnsEMBL
– Joint project between EMBL-EBI and the Sanger Centre to develop a
system which produces and maintains automatic annotation on
eukaryotic genomes.
• The SNP Consortium
– Information about up to 300000 SNPs.
• Many more…
11
Linkage Disequilibrium (LD)
• LD measures the correlation between two
SNPs.
– Some combinations of alleles or genetic
markers occur more or less frequently in a
population than would be expected from a
random formation of haplotypes from alleles
based on their frequencies.
– Non-random associations between genes at
different loci are measured by the degree of
linkage disequilibrium (D).
– Consider two loci case (i.e. two SNPs)
• SNP1 has alleles A, a
• SNP2 has alleles B, b
– When the two loci are independent, expected
freq of haplotype AB is pAB = pApB
– LD measure: D = pAB - pApB
Haplotype Frequency
A
a
B
pAB paB
b
pAb
pab
Allele Frequency
A
pA=pAB+pAb
a
pa=paB+pab
B
pB=pAB+paB
b
pb=pAb+pab
12
LD measures
• D
D = pAB – pApB, pAB = pApB + D
pAb = pA – pAB = pA – pApB – D = pA(1-pB) – D = pApb – D
A
a
Total
B
pAB = pApB + D
paB = papB − D pB
b
pAb= pApb − D
pab = papb +D
pb
Total
pA
pa
1
• D’ = D/Dmax
• r2 = D/(pApapBpb)
13
Types of Diseases
Monogenic & Complex Diseases
• Monogenic diseases – rarer (<0.1%)
– Mutated gene is entirely responsible for the disease
– Easy to locate diseased gene using LD based association studies.
• Complex diseases (more common)
– Interaction of multiple genes in a complicate fashion
• One mutation does not cause disease
• Hard to analyze – a single SNP may show weak
association
• A specific combination may show strong association, but
what combination ?
– Multiple independent causes
• There are different causes and each of these causes can
be result of interaction of several genes
• Each cause explains a certain percentage of cases
14
Tag SNP selection
15
Tag SNP
• SNPs are inherited from one generation to another in
blocks.
• Each block contains a few common haplotypes and the
SNPs in the block are in LD.
• Because of LD, each block contains a minimal
informative set of SNPs that can represent the rest of the
SNPs with high accuracy and also can identify all the
haplotypes of the block.
– Tag SNPs.
• Study of genetic factors for complex diseases
– Several genes contribute together to the disease.
– Need to study a relatively large number of SNPs.
• Also need a bigger sample size of individuals.
16
Tag SNP problem definition
• Genotyping a large number of SNPs is costprohibitive.
– Essential to choose a set of SNPs to be genotyped
such that this set predicts the rest of the SNPs (not
typed) with high accuracy.
– This set of SNPs is called the tag SNPs.
• Tag SNP selection deals with finding a set of tag SNPs of
minimum size that would have very good prediction ability for
the rest of the SNPs.
17
LD based tag SNP selection
• Greedy algorithm to identify subsets of tagSNPs for genotyping
• Start with all SNPs above a MAF threshold and calculate pair-wise
LD.
• Select the SNP that exceeds a LD threshold with the maximum
number of other sites.
– This maximally informative SNP and all associated SNP are grouped as
a bin of associated sites.
• All pairwise LD within bin are re-evaluated, and any SNP exceeding
threshold LD with all other sites in the bin is specified as a tagSNP
for the bin.
• Repeat the bining process analyzing all as-yet-unbinned SNPs at
each round, until all sites exceeding the MAF threshold are binned.
• If an SNP does not exceed the LD threshold with any other SNP in
the region, it is placed in a singleton bin.
18
Tag SNP using feature selection
• Given N x M matrix
– N haploid sequences
– M snps
• Each snp is a feature.
• Select the minimum set of features to classify all
haplotypes accurately.
• r2 = (pABpab – pAbpaB)/(pABpAbpaBpab)
• FSFS selects the most informative set of SNPs by first
grouping them into homogenous subsets and then
choosing a representative SNP from each group.
• Designed only for haplotypes
Phuong T. M., Lin Z., Altman R. B. Choosing SNPs Using Feature Selection. Proc IEEE Comput Syst Bioinform Conf.
2005; 301-9.
19
Feature selection algorithm
• Let, set of all SNPs : S = {F1; F2; ...;FN}.
• D(Fi; Fj) represents the dissimilarity between
the two SNPs (Fi and Fj ) and is calculated
using r2.
• R represent the final set of SNPs chosen as the tag SNPs.
• FSFS takes as input S and K (# of nearest neighbors of a SNP to
consider),
• During each iteration, FSFS calculates the distance D(i,k) between
each SNP F(i) in R and its kth nearest neighboring SNP.
• The algorithm then finds SNP F0 for which D(0,k) is minimum,
retains this SNP in R and removes its K nearest SNPs from R.
– Thus the algorithm always discards SNPs from the most compact
cluster causing the minimum information loss.
• FSFS gradually decreases K and re-computes D(0,k) until D(0,k) is
less than or equal to a threshold.
Phuong T. M., Lin Z., Altman R. B. Choosing SNPs Using Feature Selection. Proc IEEE Comput Syst Bioinform Conf.
2005; 301-9.
20
A Regression based method
• Uses Multivariate Linear Regression (MLR)
• SNP value prediction
M samples
k snps
– (n+1)x(k+1) matrix M corresponding to n sample
individuals and the individual x and k tag SNPs (assume
already known for prediction purpose) and a single nontag SNP s (whose value the tag SNPs will predict).
– All SNP values in M are known except the value of s in x.
– In case of haplotypes, there are only two possible
resolutions of s, s0 (for SNP value 0) and s1 (for SNP
value 1).
– For genotypes, there are 3 possible resolutions s0 (SNP
value 0), s1 (SNP value 1), and s2 (SNP value 2).
– The SNP prediction method should predict correct
resolution of s.
0 1 0 1… 1
1 0 0 1… 0
……………
1 1 0 1… 1
1
1 1…
s
.. 0
Jingwu H. and Zelikovsky A. Tag SNP Selection Based on Multivariate Linear Regression. Proc. of Intl Conf on Computational
Science (ICCS 2006), May 2006, LNCS 3992, pp. 750-757.
21
0
1
..
MLR
•
•
•
The set of tag SNPs T are vectors in the (n+1)-dimensional Euclidean
space.
Get the projections of the vectors s0, s1 and s2 onto the span of the set
of tag SNPs.
The most probable resolution of s should be closest to the span of T.
A Greedy Algorithm
1.
Start with selecting the best tag t0
that alone predicts all other tags with
minimum prediction error,
2.
In each iteration, continue to add tags
to the set T such that T best predicts
the remaining tags.
Jingwu H. and Zelikovsky A. Tag SNP Selection Based on Multivariate Linear Regression. Proc. of Intl Conf on Computational
Science (ICCS 2006), May 2006, LNCS 3992, pp. 750-757.
22
Other methods
•
•
•
•
Entropy based methods
Support vector machines
Bayesian methods
Principal Component analysis
Haplotype tagging using support vector machines. Granular Computing, 2006 IEEE International
Conference on. Jingwu He; Jun Zhang; Altun, G.; Zelikovsky, A.; Yanqing Zhang Page(s): 758- 761
Haplotype Block Partitioning and Tag SNP Selection Using Genotype Data and Their Applications to
Association Studies - Kui Zhang, Zhaohui S. Qin, Jun S. Liu, Ting Chen, Michael S. Waterman and Fengzhu
Sun Genome Research 14:908-916, 2004
Lin Z., Altman R. B. Finding haplotype tagging SNPs by use of principal components analysis. Am J Hum
Genet. 2004 Nov;75(5):850-61.
Hampe J., Schreiber S., Krawczak M. Entropy-based SNP selection for genetic association studies. (2003)
Hum Genet 114:36-43.
23
Haplotype analysis
24
Haplotype Estimation
• Each individual has two “copies” of each chromosome.
• At each site, each chromosome has one of two alleles (states)
denoted by 0 and 1 (0 major allele, 1 = minor allele)
0 1 1 1 0 0 1 1 0
Two haplotypes per individual
1 1 0 1 0 0 1 0 0
Merge the haplotypes
2 1 2 1 0 0 1 2 0
Genotype for the individual
HapMap Project
•NIH lead project ($100M) to find common haplotypes in the Human
population.
•Haplotyping individuals is expensive.
25
Haplotyping issues
• Biological Problem: For disease association studies, haplotype data
is more valuable than genotype data, but haplotype data is hard to
collect. Genotype data is easy to collect.
• Computational Problem: Given a set of n genotypes, determine the
original set of n haplotype pairs that generated the n genotypes.
2012
0010
1011
Genotype
1010
0011
Possible valid Haplotypes
Each genotype with k 2’s (heterozygotes) can be explained by 2k haplotypes
26
Need for haplotype inference
• Why do we want to determine haplotypes for individuals
at tightly linked SNP loci?
– Haplotypes are more powerful discriminators between cases and
controls in disease association studies.
– With haplotypes we can conduct evolutionary studies.
– Use of haplotypes in disease association studies reduces the
number of tests to be carried out, and hence the penalty for
multiple testing.
• Two aspects of the problem
– Estimate the frequencies of all possible haplotypes in the
population.
– Infer the haplotypes of all individuals in the given sample.
27
Clark’s method
• Haplotype inference by A. Clark in 1990.
• With a reasonable sample size, we expect to have some individuals
homozygous at every locus, e.g. 1—0—1, or heterozygous at just
one locus, e.g. 1—0—2.
– For the first case, unambiguously identify haplotype (1—0—1),
– From the second case, two (1—0—2 and 1—0—1) haplotypes are
present in the population.
– The algorithm begins by finding all homozygotes and single SNP
heterozygotes and tallying the resulting known haplotypes.
• For each known haplotype, check if the known haplotype can be
made from some combination of ambiguous sites from an
unresolved case.
– 1—0—1 known . So resolve 2—0—2 as (1—0—1) + (0—0—0).
• This chain of inferences is continued until either all haplotypes have
been recovered, or until no more new haplotypes can be found in
this way.
28
Hardy Weinberg Equilibrium
• Consider a SNP with two alleles A,a
– 3 possible genotypes A/A, A/a and a/a.
– pA, pa are the individual allele frequencies.
• HWE assumes that a child inherits the two alleles
independently from his parents.
• A population in which A/A occurs with probability p2A, A/a
with 2pApa and a/a with p2b is said to be in HWE.
– Under a certain set of assumptions like infinite population size,
random mating etc, the genotype frequencies stabilize.
29
Maximum Likelihood Estimation
•
•
•
•
Given a SNP with alleles M, m.
Possible genotypes are M/M, M/m, m/m.
What is the probability of seeing a M/M’s, b M/m’s and c m/m’s ?
According to HWE, probability that any one particular individual
selected is MM, Mm or mm is pM2, 2pMpm, pm2.
N 2a
pM (2 pM pm )b pm2c
p(a, b, c; pM , pm )
a, b, c
• Taking log, differentiating and setting to 0 gives the maximum
likelihood estimates
• pM = (2a+b)/2N, pm = (2c+b)/2N
30
Expectation Maximization (EM)
Data (D)
Available Data Missing Data
θ = Parameters to calculate the missing data
• E-step
– The missing data is calculated using θ. This along
with the available data forms the complete data (D).
• M-step
– θ’ = Recalculate the maximum likelihood estimates of
θ from D. Repeat E-step with θ= θ’.
31
Using EM
• Consider a 2-loci case
– Bi-allelic loci
• So possible haplotypes
– AB, Ab, aB, ab.
• We are given observed counts of each
possible genotype
– 9 possible genotypes
– AABB, AABb, AAbb, AaBB, …
• Observe that only genotype AaBb can
have more than 2 different haplotypes
BB
Bb
bb
Total
AA
10
15
5
30
Aa
10
50
13
73
aa
3
13
10
26
78
28
129
Total 23
x
1-x
x = fraction of genotype AaBb that are
32
Using EM
Parameters = pAB, pAb, paB, pab (haplotype frequencies)
• Calculate pAB etc. from given genotype frequencies.
– The allele frequencies are
• pA = (30+73/2)/129 = 0.5155
• pa = (26+73/2)/129 = 0.4845
• pB = (23+78/2)/129 = 0.4806
• pb=(28+78/2)/129 = 0.5194
– The haplotype frequencies are
• pAB=[2(10)+15+10+50x]/[129(2)]
• pAb=[15+2(5)+50(1-x)+13]/[129(2)]
• paB=[50x+3+13+28(2)]/[129(2)]
• pab=[50(1-x)+13+13+10(2)]/[129(2)]
• The problem is to estimate the 4 haplotype frequencies despite not
knowing the value of x (our missing data).
33
E-step
• E-step : obtain some initial values for the haplotype frequencies
– Assume we have simply each genotype frequency as product of the
respective allele frequencies.
• p0AB = (0.5155)(0.4806)
• p0Ab = (0.5155)(0.5194)
• p0aB = (0.4845)(0.4806)
• p0ab = (0.4845)(0.5194)
– The ‘expected’ value of x given these haplotype frequencies, is
34
M-step
• M-step : maximize the parameters (haplotype frequencies) using x0
calculated at the E-step.
– Substitute x0 into the haplotype frequencies.
•
•
•
•
p1AB = [2(10)+15+10+50x]/[129(2)] = 0.27131
p1Ab = [15+2(5)+50(1-x)+13]/[129(2)] = 0.24418
p1aB = [50x+3+13+28(2)]/[129(2)] = 0.20930
p1ab = [50(1-x)+13+13+10(2)]/[129(2)] = 0.27519
• Repeat E-step and M-step until the haplotype frequencies do not
change much.
35
Other methods
• Bayesian methods
• Combinatorial methods
• Dynamic programming
Haplotype Block Partitioning and Tag SNP Selection Using Genotype Data and Their Applications to
Association Studies
Kui Zhang, Zhaohui S. Qin, Jun S. Liu, Ting Chen, Michael S. Waterman and Fengzhu Sun
Genome Research 14:908-916, 2004
V. Bafna, D. Gusfield, G. Lancia, and S. Yooseph. Haplotyping asperfect phylogeny: A direct
approach. Technical report, UC Davis,Department of Computer Science, 2002.
Bayesian Haplotype Inference via the Dirichlet Process, Xing et. al, in Proceedings of the Second
RECOMB Satellite Workshop on Computational Methods for SNP and Haplotypes, pp. 99-112;
An Entropy-Based Statistic for Genomewide Association Studies
Jinying Zhao,Eric Boerwinkle,and Momiao Xiong
Am J Hum Genet. 2005 July; 77(1): 27–40.
36
SNP-disease association study
37
Support Vector Machines
• Given training set of instance-label pairs (xi,yi), i = 1,... , L
where xi ε Rn and y ε {1,−1}L, the (SVM) seeks solution to
the following optimization problem:
• Training vectors xi are mapped into a higher dimensional
space by the function Φ.
• SVM finds a linear separating hyper-plane with the
maximal margin in this higher dimensional space.
• C > 0 is the penalty parameter of the error term.
38
Support Vector Machines
• SVM machine for binary classification. The margin to be maximized
is w that separates the hyper-plane (shown with dotted line) from the
two classes of data.
39
• Multiple Myeloma (a type of cancer) is studied.
• The data set consists of genotypes from 3000 SNPs for 80 patients
selected so that they are evenly spaced at about 1Mb apart to give a
good overall coverage of the human genome.
• Each heterozygous SNP data is coded as 0, one homozygous is
arbitrarily coded as +1 and the other as -1.
• Entropy based feature selection
– Select the most informative top 10% SNPs from the set of 3000 SNPs.
– The entropy of a data set is given by - p log2(p) - (1 - p) log2(1 - p)
where p is the fraction of examples that belong to class predisposed.
– The information gain of the split is given by the entropy of the original
data set minus the weighted sum of entropies of the two data sets
resulting from the split, where these entropies are weighted by the
fraction of data points in each set.
– The SNP features are ranked by information gain, and the top-scoring
0% of the features are selected.
• Classification of the diseased and control cases using a leave-oneout cross validation approach yields an overall classification
accuracy of 71% which is significantly better than chance (50%).
Waddell M., Page D., Zhan F., Barlogie B. and John Shaughnessy Jr. J. Predicting Cancer Susceptibility from Single-Nucleotide
Polymorphism Data: A Case Study in Multiple Myeloma, Proceedings of BIOKDD '05, Chicago, Illinois, August 2005, Aug 2005.
40
Case/Control study
A Combinatorial approach
Given : A population of n genotypes each containing
values of m SNPs and disease status.
Disease
Status
Genotypes
Healthy genotypes (Control)
Diseased Genotypes (Case)
1
0
2
1
1
2
2
2
2
1
0
2
1
0
0
1
3
0
1
0
0
1
0
0
4
1
1
0
1
2
0
1
5
2
0
1
2
0
2
1
6
0
2
2
0
0
0
0
7
1
1
2
2
1
2
0
8
0
0
2
0
0
1
0
9
2
1
1
2
1
0
2
10
0
2
0
0
2
0
1
1
1
1
2
2
2
2
0: homozygous major allele, 1: homozygous minor allele, 2 : heterozygous allele
Disease association analysis searches for risk (resistance) factor with frequency
among case (control) individuals considerably higher than among control (case)
41
individuals.
Multi-SNP extension
Multi-SNP Combination (MSC)[1,2]
• Snp(C) : subset of given SNPs.
• MSC(C) : a specific value of Snp(C).
• Cluster(C) : subset of individuals that coincides with {Snp(C),
MSC(C)} in the given genotype data.
1 2 3 4 5 6 7 8 9 status
1 0 1 1 0 1 2 1 0 2 case
2 0 1 1 1 0 2 0 0 1 case
C = (1,2,4,5,7)
3 0 0 1 0 0 0 0 2 1 case
D(C) = (1,2,4)
4 0 1 1 1 1 2 0 0 1 case
H(C) = (5,7)
Snp(C) = (3,6) 5 0 0 1 0 1 2 1 0 2 control
6 0 1 0 0 1 1 0 0 2 control
7 0 1 1 0 1 2 0 0 2 control
MSC(C) x x 1 x x 2 x x x
present in 4 cases : 1 control
How significant is this cluster ?
[1] Combinatorial Search Methods for Multi-SNP Disease Association. Brinza et. al., 2006.
[2] Combinatorial Methods for Disease Association Search and Susceptibility Prediction. Brinza et. al., 2006.
42
P-value of MSC[1,2]
• Measured P-value
– Probability that diseased/healthy distribution among exposed
to risk factor happened by chance
– Compute by binomial distribution
•
Searching for risk factors among many SNPs requires multiple
testing adjustment of the p-value
[1] Combinatorial Search Methods for Multi-SNP Disease Association. Brinza et. al., 2006.
[2] Combinatorial Methods for Disease Association Search and Susceptibility Prediction. Brinza et. al., 2006.
43
Disease Association problem formulation
Given: Each containing values of m SNPs and disease status
Case/control study data consisting of n genotypes
Find: All Risk/Resistance factors (MSCs) with p-value below 0.05
44
Searching Approaches
Exhaustive search (ES)[1,2]
• Computationally infeasible, exponential number of combinations
• Searching for 3-SNP MSC on the sample with n genotypes and m
SNPs requires O(n3m)
•
Case-closure of a MSC C is an MSC C’, with maximum number of SNPs
with fixed values, which consists of the same set of cases and minimum
number of controls.
Efficient way for finding case-closure: Extend MSC with those SNPs that
have common values in all cases.
•
i j
i j
0
2
0
0
0
1
0
0
1
1
1
1
1
1
1
0
1
0
0
0
1
0
0
1
1
2
2
0
2
2
1
0
0
0
0
0
0
2
0
1
2
2
1
2
2
case
case
case
control
control
x x 1 x x 2 x x x
MSC
Present in 2 cases : 2 controls
Case-closure
MSC’
0
2
0
0
0
1
0
0
1
1
1
1
1
1
1
0
1
0
0
0
1
0
0
1
1
2
2
0
2
2
1
0
0
0
0
0
0
2
0
1
2
2
1
2
2
case
case
case
control
control
x x 1 x x 2 x 0 x
Present in 2 cases : 1 controls
Cluster C : subset of genotypes which share the same MSC
[1] Combinatorial Search Methods for Multi-SNP Disease Association. Brinza et. al., 2006.
[2] Combinatorial Methods for Disease Association Search and Susceptibility Prediction. Brinza et. al., 2006.
45
Combinatorial Search
Combinatorial search (CS)[1,2]
• Combinatorial Search Method (CS)
–
–
–
–
–
Searches only among case-closed MSCs
Avoids checking of clusters with small number of cases
Finds significant MSCs faster than ES
Still too slow for large data
Further speedup by reducing number of SNPs
• Indexing: compress S by extracting most informative SNPs
– Tag SNP Selection
– Apply ES/CS on selected tag snps
[1] Combinatorial Search Methods for Multi-SNP Disease Association. Brinza et. al., 2006.
[2] Combinatorial Methods for Disease Association Search and Susceptibility Prediction. Brinza et. al., 2006.
46
Discussion
• Neural networks, hidden markov models,
interaction information, linkage analysis etc.
• In general machine learning methods tend to do
better than purely combinatorial methods and
also are applicable to bigger data sets with
hundreds of SNPs.
– Scalablity
• Identifying SNPs in disease association study is
more difficult, largely depends on the population
under study and often faces the problem of
replication.
47























































![[edit] Use and importance of SNPs](http://s1.studyres.com/store/data/004266468_1-7f13e1f299772c229e6da154ec2770fe-150x150.png)