* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Biological Ontologies - Protein Information Resource
Survey
Document related concepts
History of genetic engineering wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene nomenclature wikipedia , lookup
Designer baby wikipedia , lookup
Protein moonlighting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcript
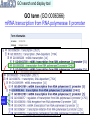
Biomedical Ontologies Bio-Trac 40 (Protein Bioinformatics) October 8, 2009 Zhang-Zhi Hu, M.D. Associate Professor Department of Oncology Department of Biochemistry and Molecular & Cellular Biology Georgetown University Medical Center 1 Overview • What is ontology? – What is biomedical ontology? • What is gene ontology? – How is it generated? – How is it used for annotation? • What is protein ontology? – Why is it necessary? – How to use it? • …… 2 Tree of Porphyry with Aristotle’s Categories Aristotle, 384 BC – 322 BC 3 Ontology: onto-, of being or existence; -logy, study. Greek origin; Latin, ontologia,1606 • In philosophy, it seeks to describe basic categories and relationships of being or existence to define entities and types of entities within its framework: – What do you know? How do you know it? – What is existence? What is a physical object? – What constitutes the identity of an object? …… • Central goal is to have a definitive and exhaustive classification of all entities. “The science of what is, of the kinds and structures of objects, properties, events, processes and relations in every area of reality” – Barry Smith, U Buffalo 4 In computer and information science • Ontology is a data model that represents a set of concepts within a domain and the relationships between those concepts. It is used to reason about the objects within that domain. Most ontologies describe individuals (instances), classes (concepts), attributes, and relations is_a Classes Relations Attributes e.g. color, engine, door… Classes (concepts) Individuals (instances) your Ford, my Ford, his Ford… 5 What are ontology useful for? Ontology is a form of knowledge representation about the world or some part of it. • • • Terminology management Integration, interoperability, and sharing of data – promote precise communication between scientists – enable information retrieval across multiple resources Knowledge reuse and decision support – extend the power of computational approaches to perform data exploration, inference, and mining Biomedical Terminology vs. Biomedical Ontology • • • • • UMLS (unified medical language system) MeSH (medical subject heading) NCI Thesaurus SNOMED / SNODENT Medical WordNet 6 Ontology Enables Large-Scale Biomedical Science The center of two major activities currently in biomedical research: • • Structured representation of biomedicine: – For different types of entities and relations to describe biomedicine (ontology content curation). Annotation: using ontologies to summarize and describe biomedical experimental results to enable: – Integration of their data with other researchers’ results – Cross-species analyses 7 Gene Ontology (GO) what makes it so wildly successful ? 8 GO Consortium http://www.geneontology.org/ • The Gene Ontology was originally constructed in 1998 by a consortium of researchers studying the genome of three model organisms: – Drosophila melanogaster (fruit fly) (FlyBase) – Mus musculus (mouse) (MGD) – Saccharomyces cerevisiae (yeast) (SGD) • Many other model organism databases have joined the GO consortium, contributing: – development of the ontologies – annotations for the genes of one or more organisms 9 Need for annotation of genome sequences • What is Gene Ontology? GO provides controlled vocabulary to describe gene and gene product attributes in any organism – how gene products behave in a cellular context Three key concepts: [Currently total 27399 GO terms] (May 2009) • Biological process: series of events accomplished by one or more ordered assemblies of molecular functions, e.g. signal transduction, or pyrimidine metabolism, and alpha-glucoside transport. [total: 16468] • Molecular function: describes activities, such as catalytic or binding activities, that occur at the molecular level. Activities that can be performed by individual gene products, or by assembled complexes of gene products; e.g. catalytic activity, transporter activity. [total: 8585] • Cellular component: a component of a cell that it is part of some larger object, maybe an anatomical structure (e.g. ER or nucleus) or a gene product group (e.g. ribosome, or a protein dimer). [total: 2346] • GO annotation - Characterization of gene products using GO terms - Members submit their data which are available at GO website. 10 GO Representation: Tree or Network? GO is a network structure root Node, a concept or a term A C Relations: is_a, or part_of C has two parents, A and B B C Leaf node 11 http://www.geneontology.org/ 12 GO search and display tool GO term (GO:0006366): mRNA transcription from RNA polymerase II promoter Leaf node 13 Human p53 – GO annotation (UniProtKB:P04637) GO:0006289:nucleotide-excision repair [PMID:7663514; evidence:IMP] 14 GO annotation of gene products • • Science basis of the GO: trained experts use the experimental observations from literature to associate GO terms with gene products (to annotate the entities represented in the gene/protein databases) Enabling data integration across databases and making them available to semantic search http://www.geneontology.org/GO.current.annotations.shtml Human, mouse, plant, worm, yeast … 15 What GO is NOT…… • Ontology of gene products: e.g. cytochrome c is not in GO, but attributes of cytochrome c are, e.g. oxidoreductase activity. • Processes, functions and component unique to mutants or diseases: e.g. oncogenesis is not a valid GO. • Protein domains or structural features. • Protein-protein interactions. • Environment, evolution and expression. • Anatomical or histological features above the level of cellular components, including cell types. Neither GO is Ontology of Genes!! – a misnomer 16 Missing GO nodes… not deep enough… not broad enough… 17 Lack of connections among GOs Estrogen receptor 18 GO: A Common Standard for Omics Data Analysis what molecular function? what biological process? what cellular component? 19 need more… • need to improve the quality of GO to support more rigorous logic-based reasoning across the data annotated in its terms • need to extend the GO by engaging ever broader community support for addition of new terms and for correction of errors • need to extend the methodology to other domains, including clinical domains, such as: – – – – – disease ontology immunology ontology symptom (phenotype) ontology clinical trial ontology ... 20 http://www.obofoundry.org/ • Establish common rules governing best practices for creating ontologies and for using these in annotations • Apply these rules to create a complete suite of orthogonal interoperable biomedical reference ontologies National Center for Biomedical Ontology (NCBO) http://bioontology.org/ 21 http://www.obofoundry.org/index.cgi?sort=domain&show=ontologies 22 The OBO Foundry • A family of interoperable gold standard biomedical reference ontologies to serve annotation of: – scientific literature – model organism databases – clinical trial data … OBO Foundry = a subset of OBO ontologies, whose developers have agreed in advance to accept a common set of principles reflecting best practice in ontology development designed to ensure: • tight connection to the biomedical basic sciences • compatibility, interoperability, common relations • support for logic-based reasoning OBO Foundry Principles: http://www.obofoundry.org/crit.shtml 23 Rationale of OBO Foundry coverage RELATION TO TIME CONTINUANT INDEPENDENT OCCURRENT DEPENDENT GRANULARITY ORGAN AND ORGANISM Organism (NCBI Taxonomy) CELL AND CELLULAR COMPONENT Cell (CL) MOLECULE Anatomical Organ Entity Function (FMA, (FMP, CPRO) Phenotypic CARO) Quality (PaTO) Cellular Cellular Component Function (FMA, GO) (GO) Molecule (ChEBI, SO, RNAO, PRO) Molecular Function (GO) Biological Process (GO) Molecular Process (GO) 24 OBO Relation Ontology Foundational is_a part_of Spatial located_in contained_in adjacent_to Temporal transformation_of derives_from preceded_by Participation has_participant has_agent e.g.: A is_a B =def. every instance of A is an instance of B “rose is_a plant all instances of rose is_a plant” 25 What is Protein Ontology? Why? http://pir.georgetown.edu/pro/ PRO 26 PRotein Ontology (PRO) PRO in OBO Foundry to represent protein entities Ontology for Protein Evolution (ProEvo) for evolutionary classes of proteins: captures the protein classes reflecting evolutionary relationships at full-length protein levels. It is meant to indicate the relatedness of proteins, not their evolution. Use is_a relationship. Ontology for Protein Forms (ProForm) for multiple protein forms of a gene: captures the different protein forms of a specific gene from genetic variations, alternative splicing, cleavage, posttranslational modifications. Relations used is_a or derives_from 27 27 Why PRO? – – – Allows specification of relationships between PRO and other ontologies, such as GO and Disease Ontology Provides a structure to support formal, computer-based inferences based on shared attributes among homologous proteins Provides a stable unique identifier to any protein type PRO:000000650 Smad2 isoform 1 phosphorylated 1 (phosphorylated SSxS motif) NOT has_function GO:0003677:DNA binding PRO:000000656 Smad2 isoform 2 phosphorylated 1 (phosphorylated SSxS motif) has_function GO:0003677 DNA binding Provides formalization and precise annotation of specific protein forms/ classes, allowing accurate and consistent data mapping, integration and analysis: for dendritic cell ontology or pathway [Term] http://www.biomedcentral.com/1471-2105/10/70 id: DC_CL:0000003 name: conventional dendritic cell def: "conventional dendritic cell is_a leukocyte that has_high_plasma_membrane_amount_relative_to_leukocyte CD11c and lacks_plasma_membrane_part CD19, CD3, C34, and CD56." [AMM:amm] comment: Immunological Reviews 2007 219: 118-142 intersection_of: CL:0000738 ! leukocyte intersection_of: has_high_plasma_membrane_amount_relative_to_leukocyte PRO:000001013 ! CD11c intersection_of: lacks_plasma_membrane_part DC_CL:0000072 ! CD3 intersection_of: lacks_plasma_membrane_part PRO:000001002 ! CD19 28 intersection_of: lacks_plasma_membrane_part PRO:000001003 ! CD34 intersection_of: lacks_plasma_membrane_part PRO:000001024 ! CD56 TGF-beta signaling – comparison between PID and Reactome PRO:000000397 LAP TGF-b Furin Ca2+ Growth signals PRO:000000616 PRO:000000410 TGF-b S P Y P Y P S P Y P T P T P MEKK1 ERK1/2 PRO:000000618 TGF-beta II I receptor PRO:000000523 S P S P T P Smad 2 Growth signals TGF-b S P Y P Y P S P Y P Stress signals II I S P Cytoplasm S P T P PRO:000000468 PRO:000000650 S SP P Smad 2 PRO:000000481 Smad 4 S P S P Smad 2 X P PRO:000000366 Shc XIAP TAK1 TPTP SPSP Smad 2 S P S P CaM Smad 2 PRO:000000651 X P Shc S P S P Smad 4 S P Y P K U TAK1 PRO:000000650 PRO:000000366 Degradation MAPKKK S P S P P38 MAPK pathway JNK cascade Smad 2 PRO:000000652 X S P S P PRO:000000650 PRO:000000366 S P S P Smad 2 Smad 2 Smad 4 Smad 4 Ski X Nucleus DNA binding and transcription regulation S P T P Y P Phosphorylation (P) at Serine (S), K U Ubiquitination (U) at Lysine (K) Threonine (T) Tyrosine (Y) Common in both Reactome & PID Only included in Reactome 29 in * All others are in PID. Not all components the pathway from both databases are listed The Need for Representation of Various Proteins Forms Glucocorticoid receptor (GR) Human PRLR and PTMs… 30 Sphingomyelin phosphodiesterase (SMPD1) (ASM_HUMAN) • Cleavage sites: – lysosomal: the enzyme is transported from the Golgi apparatus to the lysosome after additions of mannose-6-phosphate moieties (M6P) and binding to M6P receptor. – secreted: the shorter cleaved form is not modified with M6P and is targeted for secretion to the extracellular space, with different functions such as LDL binding and oxidized LDL catabolism. lysosome M6P Extracelluar, e.g. LDL binding 31 GOA for Transcription factor Ovo-like 2 Form 1 - long: GO:0045892 IDA - negative regulation of transcription, DNA-dependent Form 2 – short: GO:0045893 IDA - positive regulation of transcription, DNA-dependent - Gene. 2004 336:47-58. PMID:15225875 274 aa OVOL2_MOUSE (Q8CIV7) 32 Implications of Protein Evolution • Conclusions from experiments performed on proteins from one organism are often applicable to the homologous protein from another organism. • Information learned about existing proteins allows us to infer the properties of ancestral proteins. Common ancestor 33 Functional convergence • Protein classes of the same function derived from different evolutionary origins, e.g. carbonate dehydratase (or carbonic anhydrase EC 4.2.1.1), which has three independent gene families with functional convergence. Animal and prokaryotic type Plant and prokaryotic type Archaea type 34 Functional divergence Gene Duplication (TGM3/EPB42 split) TGM3 branch Speciation (Human/mouse split) Human TGM3 (Human) Mouse TGM3 (Mouse) Human EPB42 (Human) EPB42 branch Mouse EPB42 (Mouse) TGM3 (Human) TGM3 (Mouse) EPB42 (Human) EPB42 (Mouse) TGM3 = Protein-glutamine gamma-glutamyltransferase (Transglutaminase; involved in protein modification) EBP42 = Erythrocyte membrane protein band 4.2 (Constituent of cytoskeleton; involved in cell shape) 35 Protein Ontology (PRO) http://pir.georgetown.edu/pro/ Pfam PRO protein Root Level Domain protein domain is_a has_part GO Family-Level Distinction • Derivation: common ancestor • Source: PIRSF family translation product of an evolutionarily-related gene Gene Ontology molecular function is_a has_function biological process Gene-Level Distinction • Derivation: specific gene • Sources: PIRSF subfamily, Panther subfamily translation product of a specific gene participates_in is_a cellular component Sequence-Level Distinction • Derivation: specific allele or splice variant • Source: UniProtKB translation product of a specific mRNA part_of (for complexes) (for compartments) located_in OMIM Disease derives_from Modification-Level Distinction • Derived from post-translational modification • Source: UniProtKB Modification Level ProForm Sequence Level Gene Level ProEvo Family Level Root Level cleaved/modified translation product Example: TGF-beta receptor phosphorylated smad2 isoform1 is a phosphorylated smad2 isoform1 derives_from smad2 isoform 1 is a smad2 is a TGF-b receptor-regulated smad is a smad is a protein disease agent_in SO Sequence Ontology sequence change has_agent (sequence change) agent_of (effect on function) PSI-MOD Modification protein modification has_modification 36 Mothers against decapentaplegic homolog 2 Smad 2 GO annotation of SMAD2_HUMAN: Cellular Component: - nucleus - cytoplasm Molecular Function: - protein binding Biological Process: - signal transduction - regulation of transcription, DNA-dependent 37 TGF-b TGF-beta receptor II I Smad 2 1 phosphorylation CAMK2 ERK1 P Smad 2 Smad 2 P Smad 4 P 2 complex formation P P P P Smad 2 P P P Smad 4 3 nuclear translocation Cytoplasm P Nucleus Smad 2 P P Smad 4 4 DNA binding Transcription Regulation ++ 38 Smad2 gene products Forms Location “normal” •Cytoplasmic PRO:00000011 SMAD2_HUMAN Smad 2 Smad 2 P Smad 2 P P Smad 2 P Smad 2 Smad 2 P Smad 2 ID x P P P P TGF-b receptor phosphorylated •Forms complex •Nuclear •Txn upregulation PRO:00000013 SMAD2_HUMAN ERK1 phosphorylated •Forms complex •Nuclear •Txn upregulation++ PRO:00000014 SMAD2_HUMAN CAMK2 phosphorylated •Forms complex •Cytoplasmic •No Txn upregulation PRO:00000015 SMAD2_HUMAN alternatively spliced short form •Cytoplasmic phosphorylated short form •Nuclear •Txn upregulation point mutation (causative agent: large intestine carcinoma) •Doesn’t form complex •Cytoplasmic •No Txn upregulation P PRO:00000016 SMAD2_HUMAN PRO:00000018 SMAD2_HUMAN PRO:00000019 SMAD2_HUMAN 39 Search:smad2 -> 21 protein terms http://pir.georgetown.edu/cgi-bin/pro/textsearch_pro 40 PRO hierarchy for smad2 isoform 2 Root:Protein 41 42 PRO entry report shows the ontology and the annotation 43 44 Summary • • • • The vision of the biomedical ontology community is that all biomedical knowledge and data are disseminated on the Internet using principled ontologies, such that they are semantically interoperable and useful for improving biomedical science and clinical care. The scope extends to all knowledge and data that is relevant to the understanding or improvement of human biology and health. Knowledge and data are semantically interoperable when they enable predictable, meaningful, computation across knowledge sources developed independently to meet diverse needs. Principled ontologies are ones that follow NCBO-recommended formats and methodologies for ontology development, maintenance, and use. 45