* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Could distal MSH2 upstream deletions cause HNPCC?
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Designer baby wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Primary transcript wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
X-inactivation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene therapy wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Genome (book) wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
DiGeorge syndrome wikipedia , lookup
Alternative splicing wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Frameshift mutation wikipedia , lookup
Point mutation wikipedia , lookup
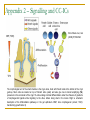
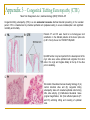
Could distal MSH2 upstream deletions cause HNPCC? … another cancer susceptibility factor. John Taylor ([email protected]) CMGS Spring Conference 2009 Hereditary non-polyposis colorectal cancer. HNPCC (Lynch syndrome) Mutations in the Mismatch repair (MMR) pathway. Four genes MLH1 PMS2 MSH2 MSH6 Colorectal cancer (right hand side). Extra-colonic carcinomas (MSH2 & MSH6). Endometrial Stomach Small bowel Hepatobiliary tract Hereditary non-polyposis colorectal cancer. U A T G T A T MSH2 MSH6 G C G C Patient referrals. “A” Patient Initials Previous molecular analysis “Z” MSH2 / MLH1 –ve mutation screen MSH2 / MLH1 –ve mutation screen IHC analysis Loss of MSH2 Loss of MSH2 MSI MSI-H MSI-H Other affected family members Mother [“B”] None n a l Interrogation of the MSH6 gene. 4 0 0 0 0 A i g Patients “A” and “Z” were referred for an MSH6 mutation screen. S 3 5 0 0 0 3 0 0 0 0 y e 2 5 0 0 0 2 0 0 0 0 D 1 5 0 0 0 1 0 0 0 0 5 0 0 0 Direct sequence analysis . MLPA analysis (MRC Holland kit P008) 1 0 0 1 5 0 2 0 0 2 5 0 3 0 0 3 5 0 4 0 0 4 5 0 5 0 0 S iz e ( n t) n a l 0 i g 4 5 0 0 0 4 0 0 0 0 Both patients were normal and had no mutations other than neutral polymorphisms in MSH6 and no deletions were identified. S 3 0 0 0 0 e B 3 5 0 0 0 y 2 5 0 0 0 D 2 0 0 0 0 1 5 0 0 0 1 0 0 0 0 5 0 0 0 0 1 0 0 1 5 0 2 0 0 2 5 0 3 0 0 3 5 0 4 0 0 4 5 0 5 0 0 a l S iz e ( n t) n 4 5 0 0 0 i g 4 0 0 0 0 C 3 5 0 0 0 S However, the MLPA kit contained additional probes: 3 0 0 0 0 y e 2 5 0 0 0 2 0 0 0 0 D 1 5 0 0 0 1 0 0 0 0 5 0 0 0 TACSTD1 exon 7 TACSTD1 exon 9 0 1 0 0 1 5 0 2 0 0 2 5 0 3 0 0 3 5 0 4 0 0 4 5 0 5 0 0 S iz e ( n t) Figure 3.1. Multiplex ligation-dependant probe amplification (MLPA) electropherogram traces for; A) a normal control; B) patient “A”; C) patient “Z”. SNPs excluded by direct sequence analysis of exon 9 TACSTD1 exon 9 deletion. This region chromosome 2p is very repetitive; Alu-rich sequence. Previous studies have shown deletions within the coding region of MSH2 can extend into the promoter region and as far as the TACSTD1 gene. Francoise Charbonnier et al., (2005) Human Mutation 26(3), 255-261. Recombination between Alu elements have also been shown (ex1-ex6). Anja Wager et al., (2003) Am J. Hum. Genet.72:1088-1100. Could the apparent deletion in TACSTD1 ex9 extend into the promoter of MSH2? QF-PCR analysis. Charbonnier et al., (2002) Cancer Research 62: 848-853. MSH2 start - 23.4 kb - 16.5 kb Ex 7 P6 P5 + 6.9 kb P4 PZ P3 PH1 TACSTD1 exon 7 TACSTD1 exon 8 TACSTD1 exon 9 P2 P1 Ex 1 MSH2 exon 1 Ex 3 MSH2 exon 2 MSH2 promoter ~4.4Kb* Msh2 Ex 6 Normal Msh2 Ex 6 Msh2 Ex 6 Msh2 Ex 16 “Z” Msh2 Ex 6 Msh2 Ex 6 “A” Map of the exons within chromosome 2 (positions 47,483,612 - 47,642,954) *promoter region as determined by Iwahashi Y et al.,(1998) Gene 213:141–147. MSH2 exon 3 QF-PCR analysis. Charbonnier et al., cancer Research 62, 848-853 MSH2 start - 23.4 kb - 16.5 kb Ex 7 P6 P5 + 6.9 kb P4 PZ P3 PH1 TACSTD1 exon 7 TACSTD1 exon 8 TACSTD1 exon 9 P2 P1 Ex 1 MSH2 exon 1 Ex 3 MSH2 exon 2 MSH2 promoter ~4.4Kb* Msh2 Ex 6 Msh2 Ex 6 Msh2 Ex 6 Msh2 Ex 16 Msh2 Ex 6 Msh2 Ex 6 “A” “B” “Z” Map of the exons within chromosome 2 (positions 47,483,612 - 47,642,954) *promoter region as determined by Iwahashi Y et al.,(1998) Gene 213:141–147. MSH2 exon 3 Long-Range PCR. Standard curve using λ DNA ladder 0 λ KB 23.1 9.4 6.6 2.3 2 1 2 3 4 5 6 7 8 0 0.2 0.4 0.6 0.8 1 1.2 1.4 1.6 1.8 2 9 10 y = -0.1821x + 1.8923 R2 = 0.9669 Series1 Series2 Linear (Series2) Linear (Series1) NC Z A B -VE sample nc "Z" 1 "Z" 2 "A" 1 "A" 2 "B" 1 "B" 2 distance (cm) size estimation lambda ladder 4.2 4.2 5.7 4.2 4.45 4.2 4.45 1.12748 1.12748 0.85433 1.12748 1.081955 1.12748 1.081955 The primer set for MSH2P4 is 10 kb upstream of MSH2 estimated size in Kb 13.4 13.4 7.2 13.4 12.1 13.4 12.1 What was the significance of these deletions? Long-range PCR excluded the possibility of rearrangements and inversions within this region. chromosomal All deletions are in excess of 10kb upstream of the MSH2 transcriptional start and are unlikely to affect transcription factor binding sites. Farré et al., (2007) Genome Biology 8:R140. The deletion co-segregates with the apparent loss of MSH2 by IHC (although only through one generation) Two additional patients from the Salisbury lab were tested and found to have the same deletion size and haplotype (two markers [D2S123 and D2S391]) as patients “A” and “B”. Findings up until Nov 2008 Recent publications MSH2 start - 23.4 kb - 16.5 kb Ex 7 P6 P5 + 6.9 kb P4 PZ P3 PH1 TACSTD1 exon 7 TACSTD1 exon 8 TACSTD1 exon 9 Publications since January 2009: P2 P1 Ex 1 MSH2 exon 1 Ex 3 MSH2 exon 2 MSH2 exon 3 MSH2 promoter ~4.4Kb* 1. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Ligtenberg et al., 2009. Nat. Gen. Vol 41 (1) p112-117. 2. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to lynch syndrome. Kovacs et al., 2009. Hum. Mut. Vol. 30, No. 2, 197–203. Both publications agree that the removal of the TACSTD1 polyadenylation signal in exon 9 resulted in the continuation of the transcriptional elongation from the TACSTD1 gene into the downstream MSH2 gene. Concluding remarks. What started of as a coincidental finding appears to a novel cancer susceptibility factor associated with HNPCC. Upstream deletions within exon 9 of the TACSTD1 gene can lead to the loss of MSH2 in colonic adenoma tissue, detectable by IHC, and is consistent with a diagnosis of HNPCC. It is unlikely that these deletions alter the expression on MSH2 directly (i.e. removing transcription factor binding sites), but rather indirectly by transcriptional silencing due to the strength of the TACSTD1 promoter and loss of the polyadenylation signal. One further consideration: Could this type of mutation influence the spectrum of cancers associated with MSH2 traditionally associated with extra-colonic carcinomas? Acknowledgements. Jennie Bell Fiona Macdonald Matthew Smith Dave Bunyan (Salisbury) Appendix 1 – Functions of TACSTD1/EpCAM Trzpis et al., (2007) The American Journal of Pathology, Vol. 171, p386-395 Appendix 2 – Signalling and CC-ICs Ricci-Vitiani et al., Gut (2008);57;538-548 The morphological unit of the small intestine is the crypt–villus, lined with Paneth cells at the bottom of the crypt (yellow). Stem cells are located on top of Paneth cells (violet) and also give rise to transit amplifying (TA) precursors in the remainder of the crypt. TA cells undergo terminal differentiation under the influence of gradients of morphogenetic ligands while migrating to the villus, before being shed in the lumen. Right: a schematic description of the differentiation pathways in the gut epithelium. BMP, bone morphogeneic protein; TGFβ, transforming growth factor β. Appendix 3 - Congenital Tufting Enteropathy (CTE) Taken from Sivagnanam et al., Gastroenterology (2008);135:429–437 Congenital tufting enteropathy (CTE) is a rare autosomal recessive diarrheal disorder presenting in the neonatal period. CTE is characterized by intestinal epithelial cell dysplasia leading to severe malabsorption and significant morbidity and mortality. Patients P1 and P2 were found to be homozygous G>A substitution in the affected patients at the donor splice site (c.491+1G>A) of exon 4 of TACSDT1/EpCAM EpCAM function may be important for the development of the crypt villus axis, where epithelial cells originate from stem cells in the crypt and migrate distally to the tip of the villus prior to shedding. Schematic of duodenal mucosa showing histology of (A) normal intestinal villus and (B) congenital tufting enteropathy villus with crowded epithelial cells forming tufts, villus atrophy. (C) H&E-stained duodenal tissue (original magnification, 20x )from affected patients (P1 and P2) exhibiting tufting and crowding of epithelial cells.