* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene Ontology and Annotation
Gene therapy wikipedia , lookup
Metagenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Pathogenomics wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene nomenclature wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene desert wikipedia , lookup
Genome (book) wikipedia , lookup
Genome evolution wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
8/24/2016
CSI/BINF 5330, Advanced Computational Biology
Gene Ontology and Annotation
Young-Rae Cho
Assistant Professor
Department of Computer Science
Baylor University
Ontology
Ontology in Philosophy
The study of the nature of being or existence including their categories
and their relations (wikipedia)
Ontology in Computer Science
The specification of a conceptualization: description of the concepts and
relationships that exist for an agent or a community of agents (Gruber)
A set of representational primitives (i.e., classes, attributes, and
relationships) for modeling a domain of knowledge
Ontology in Biology and Bioinformatics
A formal way of representing biological knowledge which is described by
the concepts and their relationships to each other (Bard and Rhee)
1
8/24/2016
Representation of Ontology
Components
Concepts and Relationships
Representation
Graph (concepts nodes, relationships edges)
Tree
Directed Acyclic Graph (DAG)
Directed Graph
Relationships in Ontology
Directions
Relationships are generally directed
Concepts have parent-child relationships
Properties (in tree or DAG)
Antisymmetric
Transitivity
Examples
“is-a” relationship
“part-of” relationship
2
8/24/2016
MIPS Functional Catalogue
Features
The functional concepts are nodes
Tree structure for the relationships between functional concepts
Provides functional annotation for small model organisms
(functional descriptions of genes or proteins)
Manually created
Well-suited for the annotation of genome from different domains of life
http://mips.helmholtz-muenchen.de/funcatDB/
Gene Ontology (GO)
Features
Organized by GO Consortium
A repository of bio-ontology (controlled vocabularies) databases
The GO terms are nodes
DAG for the relationships between GO terms
Structured in 3 main categories: biological processes, molecular functions,
- consistent descriptions across different organisms
and cellular components
Provides annotation of genes and gene products
Created by any published evidence (mostly, from high-throughput data)
Data curation, e.g., redundant annotation elimination
http://www.geneontology.org/index.shtml
3
8/24/2016
GO Structure
Example of GO DAG structure
Research Topic 1. Semantic Similarity Analysis
Definition of Semantic Similarity
Ontological relatedness between two concepts
In Gene Ontology, similarity between two terms
Categories
Ontology structure-based methods
•
Edge-based methods
•
Node-based methods
Annotation-based methods
Integrative methods
4
8/24/2016
Edge-Based Measures
Path length between two terms
Normalized path length between two terms by GO depth
Depth to the most specific common ancestor
Normalized depth to the most specific common ancestor by average depth to
two terms
where C0 is the most specific common ancestor term
Node-Based Measures
Number of common ancestors
where Pt(C) is the set of ancestors of the term C
Normalized number of common ancestors
Jaccard index
Dice index
Min normalization
5
8/24/2016
Information Contents
Formulation
In Information Theory, the information content of a concept C is defined
as -log P(C)
Transitivity Property of Annotations
If a gene g is annotated to a term C, then it is also annotated to all
the ancestor terms of C towards the root
The likelihood of C can be defined by the annotation on C
P(C) =
the number of genes annotated to C
the number of all genes annotated to the ontology
Annotation-Based Measures
Information content of the most specific common ancestor
where C0 is the most specific common ancestor
Normalized information content of the most specific common ancestor by
average information content of two terms
Sum of differences between information content of the most specific common
ancestor and information content of two terms
6
8/24/2016
Integrative Methods
Combination of an edge-based measure and a node-based measure
Combination of a node-based measure and an annotation-based measure
Combination of two annotation-based measures
Problems of Semantic Similarity
Node-Based Methods
Assumes that all GO terms are meaningful
(Terms have been randomly created based on evidence.)
Edge-Based Methods
Assumes that all relationships represents the same quantity of similarity
(Relationships have been randomly created based on evidence.)
Annotation-Based Methods
Applicable only if genes are fully annotated
7
8/24/2016
Applications of Semantic Similarity
Applications
Functional prediction of incompletely annotating genes
Semantic similarity between terms (concepts)
Functional similarity between genes
Challenges
A single gene performs multiple functions
A single gene is annotated on multiple terms
•
X={X1, X2, … , Xm} are the most specific terms having a gene g1
•
Y={Y1, Y2, … , Yn} are the most specific terms having a gene g2
Conversion to Functional Similarity
Pairwise Averaging
Average of semantic similarity scores between Xi and Yi
Best Matching
Maximum semantic similarity score between Xi and Yi
Best-Match Averaging
8
8/24/2016
Research Topic 2. Association Rule Analysis
Definition of Association Rules
One-directional relationship between two sets of items
In Gene Ontology, association rules from a term A to a term B
Categories
Pairwise association rules vs. Multi-terms association rules
Within-ontology rules vs. Cross-ontology rules
Association Rules Basics (1)
Background
Used to solve a market basket problem
Found from a transaction database
•
Transaction: a set of items that are bought by one person at one time
•
Transaction database example
T‐ID
Items
1
bread, eggs, milk, diapers
2
coke, beer, nuts, diapers
3
eggs, juice, beer, nuts
4
milk, beer, nuts, diapers
5
milk, beer, diapers
9
8/24/2016
Association Rules Basics (2)
Frequent itemsets
A set if items (as a subset of any single transaction) which occur
Itemset having support greater than (or equal to) a user-specified
frequently across transaction
minimum support threshold
Support
Frequency of a set of items across transactions
support (A → B) = P(A U B)
Association Rules Basics (3)
Association Rules
One-directional relationship between two sets of items (e.g., A → B)
Rules having confidence greater than (or equal to) a user-specified
minimum confidence threshold
Confidence
For A → B, percentage of transactions containing A that also contain B
confidence (A → B) = P(B | A) =
P (A U B)
P (A)
10
8/24/2016
Association Rule Mining
Process
(1) Find frequent itemsets
(2) Find association rules
Brute Force Algorithm
Enumerate all possible subsets of the total itemset
Count frequency of each subset
Select frequent itemsets
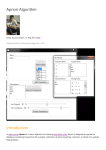
Apriori Algorithm
Iterative increment of the itemset size
(1) Candidate itemset generation
(2) Frequent itemset generation
Apriori Algorithm Details
Downward Closure Property
Any superset of an itemset X cannot have higher support than X
If an itemset X is frequent (support of X is higher than minimum support),
then any subset of X must be frequent
Candidate Itemset Generation
Selective joining
•
Each candidate itemset with size k is generated by joining two
•
The frequent itemsets with size (k-1) which share a frequent sub-
frequent itemsets with size (k-1)
itemset with size (k-2) are joined
A priori pruning
•
A frequent itemset with size k which has any infrequent sub-itemsets
with size (k-1) is pruned
11
8/24/2016
Applications of Association Rules
Applications
Application to GO annotation data
Gene ID
GO terms
1
Term‐1, Term‐4, Term‐6
2
Term‐2, Term‐4
3
Term‐1, Term‐3, Term‐4
4
Term‐3, Term‐5, Term‐6, Term‐7
Curation of GO (by within-ontology rules)
Functional prediction of of incompletely annotating genes
Correction of inconsistent annotations
Questions?
Lecture Slides are found on the Course Website,
web.ecs.baylor.edu/faculty/cho/5330
12