* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 2.2
Survey
Document related concepts
History of virology wikipedia , lookup
Hospital-acquired infection wikipedia , lookup
Quorum sensing wikipedia , lookup
Horizontal gene transfer wikipedia , lookup
Microorganism wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Phospholipid-derived fatty acids wikipedia , lookup
Human microbiota wikipedia , lookup
Triclocarban wikipedia , lookup
Disinfectant wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Marine microorganism wikipedia , lookup
Transcript
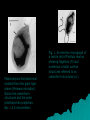
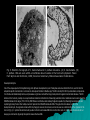
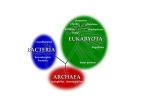
POKOK BAHASAN 1. PENDAHULUAN 2. PENGGOLONGAN MIKROORGANISME 3. STRUKTUR DAN FUNGSI SEL MIKROORGANISME 4. PERTUMBUHAN MIKROORGANISME 5. GENETIKA MIKROORGANISME 6. BIOENERGETIKA MIKROORGANISME 7. INTERAKSI MIKROORGANISME 8. PENYEBARAN MIKROORGANISME 9. PENGENDALIAN PERTUMBUHAN MIKROORGANISME 10.PERANAN MIKROORGANISME MIKROBIOLOGI DASAR 02. PENGGOLONGAN MIKROORGANISME POKOK BAHASAN I. PENDAHULUAN II. EVOLUSI DAN KERAGAMAN MIKROBA III. TINGKATAN TAKSONOMI IV. SISTEM KLASIFIKASI V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI VI. FILOGENI MIKROBA VII. DIVISI UTAMA ORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI A. Karakter Klasik 1. Karakter morfologis Mudah analisisnya, genetik stabil, dan variasi tidak dipengaruhi lingkungan; seringkali mengindikasikan hubungan filogenetik 2. Karakter fisiologis dan metabolik Secara langsung berkaitan dengan enzim dan protein transport (produk gen) dan oleh karenanya memberikan perbandingan langsung genom mikroba 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI A. Karakter Klasik (lanjutan) 3. Karakter ekologis Meliputi pola siklus hidup, hubungan simbiotik, kemampuan menyebabkan penyakit, preferensi habitat, dan kebutuhan hidup 4. Analisis genetik Meliputi studi pertukaran gen kromosomal melalui transformasi dan konjugasi; Jarang terjadi antar genera Harus dihindari kesalahan yang disebabkan sifat yang berasal plasmid 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI B. Karakter Molekuler 1. Komparasi protein – dapat memberikan informasi genetik organisme Caranya: • Penentuan urutan asam amino protein • Komparasi mobilitas elektroforesis • Penentuan reaktifitas-silang imunologis • Komparasi sifat enzimatis 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI B. Karakter Molekuler (lanjutan) 2. Komposisi basa asam nukleat a. Kandungan G+C dapat ditentukan dari suhu leleh (melting temperature, Tm). Tm adalah suhu di mana dua untai molekul DNA memisah datu sama lain ketika suhu dinaikkan perlahan-lahan. b. Secara taksonomis berguna karena variasi di dalam suatu genus biasanya kurang dari 10% tetapi variasi di antara genera cukup besar, berkisar antara 25 – 80%. 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI B. Karakter Molekuler (lanjutan) 3. Hibridisasi Asam Nukleat a. Penentuan tingkat homologi urutan b. Suhu inkubasi mengendalikan tingkat homologi urutan yang diperlukan untuk membentuk hibrid stabil 02. PENGGOLONGAN MIKROORGANISME V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI B. Karakter Molekuler (lanjutan) 4. Sekuensing Asam Nukleat a. Urutan gen rRNA adalah sangat ideal untuk komparasi karena mengandung urutan yang stabil (lestari) dan bervariasi evolusioner b. Akhir-akhir ini, genom prokariotik lengkap telah disekuensing; komparasi langsung urutan genom lengkap tidak diragukan lagi berperan penting dalam taksonomi prokariotik 02. PENGGOLONGAN MIKROORGANISME POKOK BAHASAN I. PENDAHULUAN II. EVOLUSI DAN KERAGAMAN MIKROBA III. TINGKATAN TAKSONOMI IV. SISTEM KLASIFIKASI V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI VI. PENDUGAAN (ASSESSING) FILOGENI MIKROBA VII. DIVISI UTAMA ORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT X. MENGENAL LEBIH DEKAT ANGGOTA DUNIA MIKROBA 02. PENGGOLONGAN MIKROORGANISME VI. PENDUGAAN FILOGENI MIKROBA Taksonomi prokatiotik berubah sangat cepat. Hal ini disebabkan: perkembangan pengetahuan biologi prokariotik kemajuan ilmu komputer Penggunaan karakter molekuler dalam menentukan hubungan filogenetik di antara kelompok prokariotik 02. PENGGOLONGAN MIKROORGANISME VI. PENDUGAAN FILOGENI MIKROBA A. Kronometer molekuler Asumsi bahwa laju perubahan genetik konstan adalah tidak benar; namun laju perubahan gen tertentu mungkin konstan 02. PENGGOLONGAN MIKROORGANISME VI. PENDUGAAN FILOGENI MIKROBA B. Phylogenetic trees 1. Made of branches that connect nodes, which represent taxonomic units such as species or genes; rooted trees provide a node that serves as the common ancestor for the organisms being analyzed 2. Developed by comparing molecular sequences and differences are expressed as evolutionary distance; organisms are then clustered to determine relatedness; alternatively, relatedness can be estimated by parsimony analysis assuming that evolutionary change occurs along the shortest pathway with the fewest changes to get from ancestor to the organism in question 02. PENGGOLONGAN MIKROORGANISME VI. PENDUGAAN FILOGENI MIKROBA C. rRNA, DNA, and proteins as indicators of phylogeny 1. Association coefficients from rRNA studies are a measure of relatedness 2. Oligonucleotide signature sequences occur in most or all members of a particular phylogenetic group and are rarely or never present in other groups even closely related ones; useful at kingdom or domain levels 3. DNA similarity studies are more effective at the species and genus level 4. Protein sequences are less affected by organism-specific differences in G+C content 5. Analyses of the three types of molecules do not always produce the same evolutionary trees 02. PENGGOLONGAN MIKROORGANISME VI. PENDUGAAN FILOGENI MIKROBA D. Polyphasic taxonomy 1. Uses a wide range of phenotypic and genotypic information to develop a taxonomic scheme 2. Techniques and information used depend on level of taxonomic resolution needed (e.g., serological techniques are good for identifying strains, but not genera or species 02. PENGGOLONGAN MIKROORGANISME POKOK BAHASAN I. PENDAHULUAN II. EVOLUSI DAN KERAGAMAN MIKROBA III. TINGKATAN TAKSONOMI IV. SISTEM KLASIFIKASI V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI VI. PENDUGAAN (ASSESSING) FILOGENI MIKROBA VII. DIVISI UTAMA ORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT X. MENGENAL LEBIH DEKAT ANGGOTA DUNIA MIKROBA 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME A. Domains 1. Woese and collaborators used rRNA studies to group all living organism into three domains a. BACTERIA-comprise the vast majority of procaryotes; cell walls contain muramic acid; membrane lipids contain ester-linked straight-chain fatty acids b. ARCHAEA-procaryotes that: lack muramic acid, have lipids with ether-linked branched aliphatic chains, lack thymidine in the T arm of tRNA molecules, have distinctive RNA polymerases, and have ribosomes with a different composition and shape than those observed in Bacteria c. EUCARYA-have a more complex membrane-delimited organelle structure 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME A. Domains (lanjutan) 2. Several different phylogenetic trees have been proposed relating the major domains and some trees do not even support a three-domain pattern 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME A. Domains (lanjutan) 3. One of the most important difficulties in constructing a tree is widespread, frequent horizontal gene transfer; a more correct tree may resemble a web or network with many lateral branches linking various trunk VII. DIVISI UTAMA ORGANISME The Three-Domain System Figure 10.1 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME B. Kingdoms 1. Whittaker’s five-kingdom system was the first to gain wide acceptance a. ANIMALIA-multicellular, nonwalled eucaryotes with ingestive nutrition b. PLANTAE-multicellular, walled eucaryotes with photoautotrophic nutrition c. FUNGI-multicellular and unicellular, walled eucaryotes with absorptive nutrition d. PROTISTA-unicellular mechanisms eucaryotes with various e. MONERA (PROCARYOTAE ) - all procaryotic organisms nutritional 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME B. Kingdoms 2. Many biologists do not accept Whittakerís system, primarily because it does not distinguish bacteria from archaea 3. A number of alternatives have been suggested, including a sixkingdom system and a two-empire, eight-kingdom system 02. PENGGOLONGAN MIKROORGANISME POKOK BAHASAN I. PENDAHULUAN II. EVOLUSI DAN KERAGAMAN MIKROBA III. TINGKATAN TAKSONOMI IV. SISTEM KLASIFIKASI V. KARAKTERISTIK UTAMA YANG DIGUNAKAN DALAM TAKSONOMI VI. PENDUGAAN (ASSESSING) FILOGENI MIKROBA VII. DIVISI UTAMA ORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT X. MENGENAL LEBIH DEKAT ANGGOTA DUNIA MIKROBA 02. PENGGOLONGAN MIKROORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY A. The First Edition of Bergey’s Manual of Systematic Bacteriologyprimarily phenetic 1. Contains 33 sections in four volumes 2. Each section contains bacteria that share a few easily determined characteristics (e.g., morphology, gram reaction, oxygen relationships) and bears a title that describes these properties or provides the vernacular names of the bacteria included 02. PENGGOLONGAN MIKROORGANISME VIII. BERGEY’S MANUAL OF SYSTEMATIC BACTERIOLOGY B. The Second Edition of Bergey’s Manual of Systematic Bacteriology 1. Largely phylogenetic rather than phenetic 2. Consists of five volumes Volume 1: The Archaea, Cyanobacteria, Phototrophs and Deeply Branching Genera Volume 2: Gram negative proteobacteria (purple bacteria) - complex group Volume 3: Gram positive bacteria with low G + C content (< 50%) Volume 4: Gram positive bacteria with high G + C content (> 50-55%) Volume 5: The Planctomycetes, Spriocheates, Fibrobacteres, Bacteroidetes, and Fusobacteria-an assortment of deeply branching phylogenetic groups that are not necessarily related to one another although all are Gram negative 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT (The Bergey’s Manual of Systematic Bacteriology. 2nd ed.) A. Volume 1: The Archaea, and Deeply Branching and Phototrophic Genera 1. ARCHAEA divided into two phyla a. Crenarchaeota diverse phylum that contains thermophilic and hyperthermophilic organisms as well as some organisms that grow in oceans at low temperatures as picoplankton b. Euryarchaeota contains primarily methanogenic and halophilic procaryotes and also thermophilic, sulfur-reducing procaryotes 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT (The Bergey’s Manual of Systematic Bacteriology. 2nd ed.) A. Volume 1: The Archaea, and Deeply Branching and Phototrophic Genera 2. Bacteria a. Aquificae-phylum containing autotrophic bacteria that use hydrogen as an energy source; most are thermophilic b. Thermatogae-phylum containing anaerobic, thermophilic fermentative, gramnegative bacteria; have unusual fatty acids c. Deinococcus-Thermusî-this phylum includes bacteria with extraordinary resistance to radiation and thermophilic organisms d. Chloroflexi-this phylum consists of bacteria often called green nonsulfur bacteria; some carry out anoxygenic photosynthesis, while others are respiratory, gliding bacteria; have unusual peptidoglycans and lack lipopolysaccharides in their outer membranes e. Cyanobacteria-a phylum consisting of oxygenic photosynthetic bacteria f. Chlorobi-this phylum contains anoxygenic photosynthetic bacteria known as the green sulfur bacteria; 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT B. Volume 2: The Proteobacteria devoted to a single phylum called Proteobacteria, which consists of a diverse array of gram-negative bacteria 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT C. Volume 3: The Low G+C Gram-Positive Bacteria devoted to a single phylum called Firmicutes; all have a G+C content 50%; with the exception of the mycoplasmas, which lack a cell wall, they are gram positive; most are heterotrophs; includes genera that produce endospores 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT D. Volume 4: The High G+C Gram-Positive Bacteria describes the phylum Actinobacteria; have G+C content 50-55%; includes filamentous bacteria (actinomycetes) and bacteria with unusual cell walls (mycobacteria) 02. PENGGOLONGAN MIKROORGANISME IX. GARIS BESAR FILOGENI DAN KERAGAMAN PROKARIOT E. Volume 5: The Planctomycetes, Spriocheates, Fibrobacteres, Bacteroidetes, and Fusobacteria an assortment of deeply branching phylogenetic groups that are not necessarily related to one another although all are Gram negative 1. Planctomycetes - this phylum contains bacteria with unusual features, including cell walls that lack peptidoglycan and cells with a membrane-enclosed nucleoid; divide by budding and produce appendages called stalks 2. Chlamydiae - this phylum contains obligate-intracellular pathogens having a unique life cycle; they lack peptidoglycan 3. Spirochaetes - a phylum composed of helically shaped bacteria with unique morphology and motility 4. Bacteroides - this phylum contains a number of ecologically significant bacteria Planctomyces-like bacterium isolated from the giant tiger prawn (Penaeus monodon). Notice the crateriform structures and the polar prostheca-like projection. Bar = 0.5 micrometers Fig. 1. An electron micrograph of a motile cell of Pirellula marina showing flagellum (fl) and numerous circular surface structures referred to as crateriform structures (cr) Fig. 3. Electron micrograph of C. trachomatis and C. psittaci inclusions. (A) C. trachomatis. (B) C. psittaci. RBs are seen within a membrane bound vesicle in the host cell cytoplasm. Taken from Wyrick and Richmond, 1989; American Veterinary Medical Association Publications. Developmental Cycle One of the unique aspects of chlamydial biology is the biphasic developmental cycle. Chlamydiae exist as two distinct life forms, each of which is adapted to specific environments in a manner not unlike spore formation in Bacillus spp. The EB is small (200–300 nm) extracellular, and spore-like. It is infectious but metabolically inactive, and possesses a rigid outer cell wall that may provide protection against environmental stresses. The EB attaches to the host cell, possibly via a receptor/adhesin interaction. Attachment is followed by penetration into a membrane-bound vesicle where the EB differentiates into the larger (700–1000 nm) RB. RBs are noninfectious and relatively fragile but capable of synthesizing macromolecules and replicating by binary fission. After multiple rounds of replication the RBs differentiate into EBs. Throughout the intracellular portion of the developmental cycle, the organisms remain within the phagocytic vacuole, the inclusion, and can be seen as a distinct entity in the cytoplasm of the host cell (Fig. 3). C. psittaci infected host cells lyse late in infection and release EBs that can infect other cells. C. trachomatis infected cells do not always lyse and must be physically disrupted to release infectious EBs. p. 402, Figure 1, illustrates representative spirochetal organisms. Note that Borrelia is the longest and has the fewest spirals per unit length and that Spirillum is the shortest, bluntest and has the sharpest ends. The different treponemes can be differentiated by the number of spirals per length. The bottom of the graphic is a representative dark-field illuminated view of what spirochetes would look like: black background and white spiral organisms Bacteroides forsythus DIFFERENT EDITIONS OF BERGEY’S MANUAL Bergey’s Manual of Determinative Bacteriology First edition, 1923: Bergey, D. H., Harrison, F. C., Breed, R. S., Hammer, B. W., Huntoon, F. M.Bergey’s Manual of Determinative Bacteriology. The Williams & Wilkins Co., Baltimore. 442 pp. Second edition, 1925: Bergey, D. H., Breed, R. S., Hammer, B. W., Huntoon, F. M., Murray, E.G. D., Harrison, F. C. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 462 pp. Third edition, 1930: Bergey, D. H., Breed, R. S., Hammer, B. W., Huntoon, F. M., Murray, E. G.D., Harrison, F. C. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 589 pp. Fourth edition, 1934: Bergey, D. H., Breed, R. S., Hammer, B. W., Huntoon, F. M., Murray, E.G. D., Harrison, F. C. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 664 pp. Fifth edition, 1939: Bergey, D. H., Breed, R. S., Murray, E. G. D., Hitchens, A. P. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 1032 pp. Sixth edition, 1948: Breed, R. S. Murray, E. G. D., Hitchens, A. P. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 1530 pp. Seventh edition, 1957: Breed, R. S. Murray, E. G. D., Smith, N. R. Bergey’s Manual of Determinative Bacteriology, Williams & Wilkins. 1094 pp. Eighth edition, 1974: Buchanan, R. E., Gibbons, N. E. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 1268 pp. Ninth edition, 1994: Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T., Williams, S. T.Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 787 pp Contents of the Bergey’s Manual of Systematic Bacteriology. 1st ed. This work is a recognized authority on bacterial taxonomy. The Manual is divided into four volumes. Each volume contains several sections, and each section contains a number of related genera. In brief, the contents of each volume is as follows: Volume I 1984. Gram-negative Bacteria of medical and commercial importance: spirochetes, spiral and curved Bacteria, Gram-negative aerobic and facultatively aerobic rods, Gram-negative obligate anaerobes, Gram-negative aerobic and anaerobic cocci, sulfate and sulfur-reducing bacteria, rickettsias and chlamydias, mycoplasmas. Volume II 1986. Gram positive Bacteria of medical and commercial importance: Gram-positive cocci, Gram-positive endospore-forming and nonsporing rods, mycobacteria, nonfilamentous actinomycetes. Volume III 1989. Remaining Gram negative Bacteria and the Archaea: phototrophic gliding, sheathed, budding and appendaged Bacteria, cyanobacteria, chemolithotrophic Bacteria; methanogens, extreme halophiles, hyperthermophiles, Thermoplasma and other Archaea. Volume IV 1989. Filamentous actinomycetes and related bacteria Contents of the Bergey’s Manual of Systematic Bacteriology. 2nd ed. Volume 1(2001) The Archaea and the deeply brancing and phototrophic Bacteria ISBN 0-387-98771-1 Volume 2 (2001) The Proteobacteria ISBN 0-387-95040-0 Volume 3 (2002) The low G + C Gram-positive Bacteria ISBN 0-387-95041-9 Volume 4 (2002) The high G + C Gram-positive Bacteria ISBN 0-387-95042-7 Volume 5 (2003) The Planctomycetes, Spriochaetes, Fibrobacteres, Bacteriodetes and Fusobacteria ISBN 0-387-95043-5 Figure 0. The Phylogenetic Tree of Life based on Comparative ssrRNA Sequencing. The Tree shows the procaryotes in two Domains, Archaea and Bacteria. At a taxonomic level, most organisms at the tips of the Archaeal branches represent a unique Order; most organisms a the tips of the bacterial branches are classified into a unique Phylum. On the Archaeal limb, the three physiological groups are evident in the names: "thermo" and "pyro" for the extreme thermophiles; "methano" for the methanogens; and "halophiles" for the extreme halophiles. The most important, best known, and diverse groups (phyla) branching off of the Bacterial limb are the Cyanobacteria, Proteobacteria and Gram positives. Figure 2. Schematic structure of peptidoglycan of layers of pneumococcal cell wall. The building blocks of glycan strands are Nacetyl glucosamine (NAG) and N-acetyl muramic acid (NAMA) connected by a beta 1,4 glycosidic linkage. The glycan layers are connected by linked peptide crossbridges covalently attached to the muramyl residue (shown as elongated rectangular boxes). The crossbridges vary in composition and in the number of amino acid as well as in the crosspeptide linkage. Currently there are studies being performed on Prochloron's photosynthetic machinery, as its morphology is unique among prokaryotes. Physically they are structurally similar to the photosynthetic apparatus of green plastids, with the thylakoids (the saclike membranes which house the chlorophyll within the cells) located in the peripheral cytoplasm of the bacteria's cells. This feature is one of the bases for the hypothesis that this bacteria is similar to the bacteria which evolved to become the first chloroplast within photosynthesizing eukaryotes. The photosynthetic pigment composition of Prochloron has similar characteristics of both cyanobacteria and chlorophytes Characteristic Archaea Bacteria Eukaryotes Predominantly multicellular No No Yes Cell contains a nucleus and other membrane bound organelles No No Yes DNA occurs in a circular form* Yes Yes No Ribosome size 70s 70s 80s Membrane lipids ester-linked** No Yes Yes Photosynthesis with chlorophyll No Yes Yes Capable of growth at temperatures greater than 80 C Yes Yes No Histone proteins present in cell Yes No Yes Methionine used as tRNA Initiator*** Yes No Yes Operons present in DNA Yes Yes No Interon present in most genes No No Yes Capping and poly-A tailing of mRNA No No Yes Gas vesicles present Yes Yes No Capable of Methanogenesis Yes No No Sensitive to chloramphenicol, kanamycin and streptomycin No Yes No Transcription factors required No Yes Yes Capable of Nitrification No Yes No Capable of Denitrification Yes Yes No Capable of Nitrogen Fixation Yes Yes No Capable of Chemolithotrophy Yes Yes No * Eukaryote DNA is linear ** Archaea membrane lipids are ether-linked *** Bacteria use Formylmethionine "Molecular Taxonomy" Basic assumptions Genes mutate randomly Many mutations are "neutral" -- no obvious disadvantage to the strain. Once a mutation is established, all progeny carry that mutation. Organisms that differ by only a few bases have diverged more recently in evolutionary time than organisms that differ by many bases. Example: four hypothetical organisms whose DNA sequence in one homologous region is known. Conclusions: A and B differ by one base substitution C and D also differ by one base substitution A and C differ by three substitutions, A and D differ by four. Represent these differences by a distance tree (Phylogenetic tree) 02. PENGGOLONGAN MIKROORGANISME VII. DIVISI UTAMA ORGANISME