* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download C tudi - DNA to Darwin
SNP genotyping wikipedia , lookup
Primary transcript wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
DNA barcoding wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Maximum parsimony (phylogenetics) wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
DNA vaccination wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Microsatellite wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Koinophilia wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
DNA supercoil wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
DNA nanotechnology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Microevolution wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Helitron (biology) wikipedia , lookup
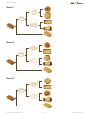
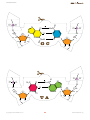
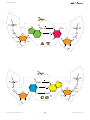
TEACHER’S GUIDE Introduction Introductory activities Dean Madden NCBE, University of Reading Version 1.0 Case Stu Case Stud introduction Introductory activities The activities in this section explain the basic principles behind the construction of phylogenetic trees, DNA structure and sequence alignment. Students are also intoduced to the Geneious software. Before carrying out the activities in the DNA to Darwin Case studies, students will need to understand: •• the basic principles behind the construction of an evolutionary tree or phylogeny; •• the basic structure of DNA and proteins; •• the reasons for and the principle of alignment; •• use of some features of the Geneious computer software (basic version). The activities in this introduction are designed to achieve this. Some of them will reinforce what students may already know; others involve new concepts. The material includes extension activities for more able students. Evolutionary trees In 1837, 12 years before the publication of On the Origin of Species, Charles Darwin famously drew an evolutionary tree in one of his notebooks. The Origin also included a diagram of an evolutionary tree — the only illustration in the book. Two years before, Darwin had written to his friend Thomas Henry Huxley, saying: ‘The time will come, I believe, though I shall not live to see it, when we shall have fairly true genealogical trees of each great kingdom of Nature.’ Today, scientists are trying to produce the ‘Tree of Life’ Darwin foresaw, using protein, DNA and RNA sequence data. Evolutionary trees are covered on pages 2–7 of the Student’s guide and in the PowerPoint and Keynote slide presentations. Page 3 of the Student’s guide is the instructions for making the tree from biscuits. DNA and protein structure Although this is well-covered in all relevant textbooks, a DNA model and animated (QuickTime) slideshow of DNA structure are provided. Sequence alignment This is shown using an animated (QuickTime) slideshow. Copyright © Dean Madden, 2011 2 www.dnadarwin.org introduction 1. Evolutionary trees This activity introduces students to the use of tree diagrams (phylogenies) to show evolutionary relationships. General reading The making of the fittest. DNA and the ultimate forensic record of evolution by Sean B. Carroll (2009) Quercus Books (Paperback) ISBN: 978 1847247247. A popular lay account of some of the molecular evidence for evolution. Reading the story in DNA: A beginner’s guide to molecular evolution by Lindell Bromham (2008) Oxford University Press (Paperback) ISBN: 978 0199290918. An engaging textbook on molecular evolution, which assumes no specialist mathematical knowledge and takes the reader from first principles. A science primer. Just the facts: A basic introduction to the science underlying NCBI resources (2004) National Center for Biotechnology Information. This document from the National Center for Biotechnology Information in the USA, provides a clear introduction to the principles of systematics and molecular phylogenetics. It can be read on-line at: http://www.ncbi.nlm.nih.gov/ About/primer/phylo.html Scientific publications These papers can be accessed free-of-charge, online. Evolution of the domestic cat Johnson, W. E. et al (2006) The late Miocene radiation of modern Felidae: A genetic assessment. Science 311, 73–77. doi: 10.1126/science.1122277 Driscoll, C. A. et al (2007) The Near Eastern origin of cat domestication. Science 317, 519–523. doi: 10.1126/science.1139518 Re-evolution of teeth in the marsupial frog, Gastrotheca guentheri Wiens, J. J. (2011) Re-evolution of lost mandibular teeth in frogs after more than 200 million years, and re-evaluating Dollo’s Law. Evolution (online advance publication). doi: 10.1111/j.1558-5646.2011.01221.x Requirements Each student or working group will need: •• cut-outs of the biscuits on page 6 of this document (these could be laminated for re-use) •• a sheet of A3 paper •• a ruler and pencil or pen •• copies of worksheets. Copyright © Dean Madden, 2011 3 www.dnadarwin.org introduction Presentations Teachers may find the PowerPoint or Keynote presentation helpful for introducing this exercise. Educational aims The activity introduces the concept of showing evolutionary relationships in a tree diagram (phylogeny). The ‘biscuit’ activity can be used as a diagnositic tool to evaluate students’ understanding of evolution, before or after teaching the subject. It emphasises how phylogenies based on phenotype alone can be misleading and introduces the concept of generating evolutionary trees based on nucleic acid and protein sequence data. The Extension material on pages 5 and 6 can be used for more able students or as homework. Prequisite knowledge Some knowledge of the basic concepts of evolution, including selection and speciation, would be useful. It is also helpful if students know the basic terms used to describe evolutionary trees e.g., root, branch, node. Classroom organisation There are several ways of using the exercise. One strategy would be to divide the class into small groups of 2–3 students and to ask each group to devise an evolutionary tree using the biscuit cut-outs. Each group then reports back to the entire class, explaining the reasons for their decisions and describing how they think the biscuits have ‘evolved’. To ensure that the discussion is meaningful, it may help to give students a list of words and phrases that they must use in their descriptions. For example: selection, speciation, convergent evolution, adaptation, mutation, common ancestor, branch, node, outgroup etc. A strategy that has proved effective is for the teacher to eavesdrop on the discussion in each group as they are devising their trees, then to choose a relatively articulate group to give their presentation first. This will then ‘set the tone’ for the other groups’ feedback. With large classes, it may prove impractical for all groups to report back, in which case just three or four groups may be asked to describe their trees verbally, then all groups can be asked to produce a written description. The exercise can also be undertaken using a selection of real biscuits, although this activity is more open-ended and hence less predictable. Health and safety regulations will have to be considered if the biscuits are to be eaten afterwards. Copyright © Dean Madden, 2011 4 www.dnadarwin.org introduction Other useful resources Simulating evolution A practical exercise in which students produce a phylogenetic tree using screws, nuts and bolts, paperclips, etc. Devised by John Barker, based on an Open University activity. Downloadable from: www.eurovolvox.org/ protocols.html Building a phylogenetic tree An exercise devised by Wojciech Grajkowski of the Science Festival School, Warsaw, in which students construct data matrices for three groups of organisms, then use them to generate phylogenies. Downloadable from: www.eurovolvox.org/protocols.html Wellcome Trust Tree of Life The Wellcome Trust Tree of Life is a six-minute animated evolutionary history presented by Sir David Attenborough. The video can be downloaded for classroom use, as can the supporting interactive tree, to be viewed using a Web browser (requires Adobe Flash 10+). An audio-free version of the video is also available for dubbing with your own soundtrack. Several additional educational resources and an animated video presentation of evolution are all free to download. www.wellcometreeoflife.org Tree of Life Web Project The Tree of Life Web Project is a collaborative effort of biologists from around the world. On more than 9,000 Web pages, the project provides information about the diversity of organisms on Earth, their evolutionary history (phylogeny), and characteristics. http://tolweb.org/tree/ Interactive Tree of Life (European Molecular Biology Laboratory) This is an advanced online tool for the display and manipulation of phylogenetic trees. It provides most of the features available in other tree viewers, and offers a novel circular tree layout, which makes it easy to visualize trees. Trees can be exported to several graphical formats, both bitmap- and vector-based. Even if you don’t want to generate your own trees, you can explore the pre-computed interactive diagrams on this site. http://itol.embl.de/ National Center for Biotechnology Information (NCBI) The NCBI hosts a major collection of databases of genetic information that is used by research scientists world-wide. The Web site includes some excellent educational materials, including a primer on systematics and molecular phylogenetics. http://www.ncbi.nlm.nih.gov/ Copyright © Dean Madden, 2011 5 www.dnadarwin.org introduction Existing (extant) species Fossil common ancestor Existing (extant) species Fossil common ancestor 6 introduction Tree 1 Tree 2 Tree 3 Copyright © Dean Madden, 2011 7 www.dnadarwin.org introduction Answers to the questions on the worksheets Page 3 There are three possible ways of constructing the evolutionary tree from biscuits. These are shown here and in the slide presentations. Page 5 a. Variations in the rate of evolution may lead to organisms being placed in the wrong place on an evolutionary tree (they may look very different when they are in fact closely-related). b. Any examples of convergent evolution could be suggested here, for example, wings in bats and birds, camera-like eyes in primates and cephalopods, streamlined body shapes in dolphins and sharks. c. For example, the genes controlling skin colour in humans have diverged very rapidly, meaning that humans look different when in fact they are all the same species. d. For evolution to go ‘in reverse’, similar selection pressures would have to apply, but once genetic diversity has been lost, the chances of successive mutations occurring to exactly recreate the original trait are remote. e. If a tree is prepared based on similarity, a re-evolved trait may cause a species to be incorrectly grouped with distantly-related organisms. f. All organisms have DNA or RNA, so there is a direct means for comparing them, which is not necessarily the case with other characteristics. Sequence data lends itself to computer-based analysis and statisitical techniques can also be applied to the data. High rates of mutation in stretches of non-coding DNA, which are not eliminated by natural selection, can be misleading however. In addition, since there are only four character states for DNA bases (C, A, G and T) the probability of any one base being shared at a particular position by chance is high. Page 6 g. i. Cat (all are mammals, the other three organisms are rodents). ii. Oak tree (the other two trees are coniferous). iii. Snail (all are molluscs, but the octopus and squid are more closely related to each other than they are to snails). iv. Wombat (all except the wombat (a marsupial) are placental mammals). v. Lemur (all are primates, but the lemur is a prosimian, while the others are simians). vi. Platypus (the kangaroo (a marsupial) and the bear (a mammal) are more closely-related to one another than they are to the egg laying platypus (a monotreme)). vii. Snail (the other two are chordates). viii. Traditionally, classification of the rosaceae (all of the species listed here) relied upon fruit type, which would imply that the strawberry is the outgroup, but genetic studies have shown that fruit type is a poor guide to evolutionary relationships here). ix. Onion (it is the only monocotyledon). x. Escherichia coli (the other two are fungi). Copyright © Dean Madden, 2011 8 www.dnadarwin.org introduction 2. A model of DNA In this activity, students make a 3-D paper model of B-DNA. There is also a QuickTime animation showing the structure of DNA. Modeling the helix This model is cheap and easy to make and shows the key features of DNA structure, including the major and minor grooves of the helix. The templates must be cut and folded with care however, and this may take a considerable time. The exercise is therefore best set as a group activity in which students prepare a few base pair templates each then combine them to build one or more models. If the nucleotide pairs are photocopied or printed onto overhead transparency sheets instead of card, these can be assembled to make a particularly attractive model. General reading The double helix. A personal account of the discovery of the structure of DNA by James D. Watson [Gunther Stent, Ed.] (1980) New York: W. W. Norton and Company. ISBN: 0 393 95075 1. What mad pursuit. A personal view of scientific discovery by Francis Crick (1988) New York: Basic Books. ISBN: 0 465 09138 5. Rosalind Franklin: The Dark Lady of DNA by Brenda Maddox (2003) London: HarperCollins. ISBN: 978 0006552116. Maurice Wilkins — The third man of the double helix by Maurice Wilkins (2005) Oxford: Oxford University Press. ISBN: 978 0192806673. Francis Crick: Discoverer of the genetic code by Matt Ridley (2008) London: HarperPerennial. ISBN: 978 0007213313. Requirements Each student or working group will need: Equipment • Scissors • Bodkin or strong needle, for punching holes through card • OPTIONAL: Sharp craft knife and cutting board Materials • Nucleotide templates, copied onto thin card (At least 16 templates will be needed to show the structure clearly) • Glue suitable for paper • Drinking straws • Fine string or strong sewing thread Copyright © Dean Madden, 2011 9 www.dnadarwin.org introduction Educational aims Knowledge of the base-pairing mechanism is essential for use of the the bioinformatics software in the DNA to Darwin Case studies. This activity can be used to introduce or revise DNA structure. Models can be made for homework or as a group activity. Prequisite knowledge Prior knowledge of the basic structure of DNA would be useful e.g., names of bases and base-pairing mechanism; sugar-phosphate backbone. Other useful resources The Wellcome Trust has several (Adobe Flash) animations of DNA structure, function and sequencing methods which can be downloaded free-ofcharge from its web site: www.wellcome.ac.uk/Education-resources/ Teaching-and-education/Animations/DNA/index.htm Fernand Schroeder of the Lycée de Garçons, Esch/Alzette (Luxemborg) has devised a cut-out ‘computer’ showing the codons that correspond to each amino acid. The reverse of the ‘computer’ shows the structure of each amino acid: www.eurovolvox.org/Animations and models/geneticcode. html The same web site also includes several interactive DNA animations devised by Tago Sarapuu and his colleagues from the Science Didactics Department, University of Tartu (Estonia). Molecule of the Month The Protein Data Bank has a tutorial on DNA structure and function, including a 3-D cut-out paper model: http://www.rcsb.org/pdb/101/motm. do?momID=23 NCBE DNA50 The NCBE has several materials on this mini web site created to celebrate 50 years of the double helix in 2003: www.ncbe.reading.ac.uk/DNA50/ menu.html Acknowledgements The model upon which this one is based was devised by Van Rensselaer Potter in 1958 and appeared the following year in his book Nucleic Acid Outlines. This article is adapted from one by the same author which first appeared in the on-line journal Bioscience Explained: www.bioscience-explained.org Copyright © Dean Madden, 2011 10 www.dnadarwin.org introduction Screenshots from the QuickTime animation 1.Use the space bar, forward or back arrow keys or mouse to move through the animation. 2.The first slide shows the components that DNA is made from and give an overview of the structure. 3.Base pairs are highlighted. Note the antiparallel arrangment of the two strands. 4.Hydrogen bonds between the bases are shown. 5.The phosphate groups are highlighted. Note the negative charge on the oxygen atoms. 6.The sugars are highlighted. Another slide shows the numbering of the carbon atoms. Copyright © Dean Madden, 2011 11 www.dnadarwin.org introduction H O CH2 O P -O N O N H H O H3C O N H N N H O N CH2 O Copyright © Dean Madden, 2011 H N GC H N O H O P O- N O N TA 12 CH2 O N O CH2 O H N N H O O H O O O P O- H N O O P O- N www.dnadarwin.org introduction CH2 O N O O H N N O O P O N N CH2 O Copyright © Dean Madden, 2011 H O O N H N 13 H CH2 O N H N CG O P O- N O N H CH2 O H H O N AT H O P O- CH3 H O N O N H O O P OO -O H N www.dnadarwin.org introduction RNA code DNA code Copyright © Dean Madden, 2011 14 www.dnadarwin.org introduction 3. Alignment This animation is useful to highlight why alignment is necessary to detect underlying relationships between nucleic acid or protein sequences. Note that inversions are not shown in the animation, as these cannot be identified by most alignment software. Generally, inverted sequences are treated by the software like any other sequence and consequently underlying relationships may not be detected. The identification of inversions in sequence data is a non-trivial and computationally-expensive task. Alignments can be produced using DNA or protein sequence data. Amino acid sequences are of course a third of the length of their nucleic acid equivalents, so they carry less information. Because of the redundancy in the Genetic Code (with one amino acid being represented by more than one codon), for a particular sequence, protein alignments show less variation than their nucleic acid equivalents. Consequently, a phylogenetic tree generated from a protein alignment may be different from that generated by the equivalent nucleic acid sequence. Requirements • Computer with QuickTime installed. QuickTime may be downloaded freeof-charge from the Apple web site: www.apple.com/quicktime • QuickTime animation showing alignment. Screenshots from the QuickTime animation 1.The animation shows three types of mutation and the alignment of four DNA sequences. Copyright © Dean Madden, 2011 2.You can use the space bar, forward or back arrow keys or mouse to move through the animation. 15 www.dnadarwin.org introduction 3.Pauses (requiring a click to advance) allow you to explain the process and to question students. 4.At the end of each section, there is a static one-screen summary of the process. 5.The three derived sequences and the original DNA sequence are aligned. 6.Note how insertions affect the alignment of all other sequences; substitutions can be seen here too. 7.An example of aligned DNA sequences in Geneious. The inserted dashes can clearly be seen. Copyright © Dean Madden, 2011 8.Aligned protein sequences. The asterisks on a black background show unknown amino acids. 16 www.dnadarwin.org