* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Quantitative Genomics slides
Vectors in gene therapy wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genetic drift wikipedia , lookup
Human genome wikipedia , lookup
X-inactivation wikipedia , lookup
Minimal genome wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression programming wikipedia , lookup
Heritability of autism wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Irving Gottesman wikipedia , lookup
Genetic testing wikipedia , lookup
Medical genetics wikipedia , lookup
Genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Human genetic variation wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Population genetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Behavioural genetics wikipedia , lookup
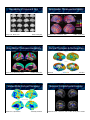
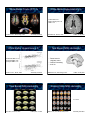
Mendelian & Complex Traits Quantitative Imaging Genomics David C. Glahn, PhD Olin Neuropsychiatry Research Center & Department of Psychiatry, Yale University Mendelian Trait Complex Trait • A trait influenced by a single gene producing a clear pattern of dominant or recessive inheritance within families. • A trait influenced by multiple genes and their interactions with each other and with the environment. • Examples: cystic fibrosis, sickle cell anemia, hemophilia • Examples: autism, schizophrenia, Alzheimer’s, brain anatomy, BOLD signal July 22, 2010 Genetics Terminology 1 Genetics Terminology 2 • Quantitative Trait Locus (QTL), a location, usually a chromosomal region, implicated as containing one or more genes that influence a phenotype of interest • Quantitative Trait Nucleotide (QTN), a specific sequence variation that has been implicated as having a functional effect on a phenotype of interest • A QTL may contain multiple QTNs • Chromosome: a single long string of DNA, humans have two copies of each chromosome (diploid), one from mom, one from dad • Autosome: None sex chromosome (22 in humans) • Sex chromosome: X or Y chromosome, females have 2X, males have 1X and 1Y • Mitochondrial DNA: non-nuclear DNA, inherited only from the mother Human Genome Genetics Terminology 3 23 Chromosomes ~20-25,000 genes ~3 billion base pairs • Gene: a unit of DNA that codes for a protein (but the term may be used to include both coding and non-coding elements) • Locus: location, sometimes used interchangably with gene • Allele: the specific variant you have at a particular site in the genome, the possible variants at a locus Loci, Alleles, Genotypes, Genetics Terminology 4 • Genotype: the combination of alleles on the two chromosomes of an individual • Haplotype: the alleles at different loci being carried together on the same chromosome • Phenotype: the trait of interest, some measurable property of the individual – Examples: neuroanatomy, neurophysiology, schizophrenia, bipolar disorder, Alzheimer’s Within A Gene A Small Gene: FVII gene (~15 kb) H115H 1 2 Two loci: letter locus & number locus Each locus has two alleles: A or B, 1 or 2 The genotype at the letter locus is AB The haplotype on the first chromosome shown is A1 • Functional & physical unit of heredity passed from parent to offspring (pieces of DNA) • Typically contain information to make a specific protein • Composed of nucleotides, sequence of four organic bases (Adenine, Guanine, Cytocine, and Thyamine) • Matching nucleotides on the complimentary DNA strands form a base-pair How to do Genetics Question A294V 9.734 A/G R353Q IVS 7 S333S 10.524 G/A -668 A/C 2 3 4 5 6 7 8 -3216 C/T -628 A/G1a 1b 10.534 T/C -2987 C/A 3.171 G/A 5.191 A/G 7.161 G/C 260 A/G 3.294 G/A 5.503 T/A 7.453 T/G 9.431 A/G 364 G/A 7.729 G/A 698 T/C 3.380 C/T 9.779 T/C 705 G/A 3.423 G/T 8.695 G/A 9.792 G/A 6.331 G/A 3.928 G/A 710 C/G 9.847 C/T 4.003 G/A 6.448 G/T 723 - 101 bp 6.452 G/T 799 T/C 6.461 + T 806 G/A 811 C/G 833T/C 945 G/A B The Gene • Exon: specifies the mRNA, which is translated into the series of amino acids in the protein, coding sequence • Intron: non-coding, intravening sequence • Splice site: the juncture between an exon & an intron • Promoter, enhancer: regulatory element (usually non-coding), controls time, place, amount of transcription -323 ± 10bp -401 G/T -122 T/C -402 G/A 73 G/A A What do you want to know? 11.912 G/A 11.622-3 del AG 11.293-4 ins AA Sample Who do you need to study? Method How will you use your data? Questions for the Study of 1) Is this trait influenced by genetic factors? How strong are these genetic influences? 2) Which traits are influenced by the same genes? 3) Where are the genes that influence a trait? 4) What are the specific genes that influence the trait? 5) What specific genetic variants influence the trait and how do they interact with each other and with the environment? Subject Ascertainment Strategies 1. By phenotype: if you’re studying a rare disease, you must ascertain on phenotype. This is also necessary for some study designs (TDT, case/control). 2. Randomly: if you’re studying a common disease, you’ll find it in a random sample. If you’re interested in multiple traits, ascertaining on one improves power only for that one. May also want to study normal variation. 3. Ascertainment also depends on (and limits) method of analysis – TDT, affected relative pair linkage. Defining Heritability Six Types of Samples for Genetics 1. Adoptees: separating the effects of genes and family environment 2. Unrelated individuals: association only, estimation of effect size after variants are identified 3. Parent-child triads: association in the presence of linkage (transmission disequilibrium test), heritability/relative risk 4. Twins: heritability, relative risk, genetic correlations, linkage, association 5. Relative pairs: heritability, relative risk, genetic correlations, linkage, association 6. Pedigrees: heritability, relative risk, genetic correlations, linkage, association Question 1: Heritability Is this trait influenced by genetic factors? How strong are these genetic influences? Variance Decomposition !"#"$% &#'()"*+#,'-.#/"#0 123-"4*+#,'-.#/"#0 ! 2 µ ^ = "x / n µ i Phenotype (P) = Genotype (G) + Environment (E) ^ ^ !2 = "(x - µ)2 / n Almasy & Blangero, Am J Hum Genet, 1998 ! ! ! ! ! ! ! ! 2 p = !g + !e 2 g = !a + !d 2 e = ! c + ! eu 2 = total phenotypic p 2 g 2 e 2 a 2 d 2 2 2 2 2 2 = genetic = environmental = additive genetic = dominance Broad-Sense Heritability (H2) • All possible genetic contributions !g 2 H= 2 !p 2 – allelic variation (additive variance) – dominance variation, – epistatic (multi-genic) – interactions, – maternal and paternal effects -a AB 0 2 2 g a d AA BB 0 d BB +a VA = 2pq[a + d(q-p)]2 +a Narrow-Sense Heritability (h2) !a h = 2 !p 2 2 AB Estimating Additive & Dominate If the heterozygote is half way between the two homozygotes, there’s a “dose-response” effect, d is zero, and there is no dominance. • Heritability (h2): the proportion of the phenotypic variance in a trait attributable to the additive effects of genes. ! = ! + ! 2 -a Defining Dominance AA Decomposing Genetic Effects VD = (2pqd)2 Conceptualizing Heritability • Heritability estimates vary between 0 and 1 – 0, genetic factors do not influence trait variance – 1, trait variance is completely under genetic control • If h2=0.5, then 50% of phenotypic variation is due to genetic variation. – Not that the trait is 50% caused by genetics • Stronger heritability does not imply simple genetics Estimating Heritability with Twins Falconer’s Method Estimating Heritability with Twins • Monozygotic (identical): • Dizygotic (fraternal): h2=2*(rMZ-rDZ) VA+VD 1/2 VA + 1/4 VD • 2 x Difference = VA + 3/2 VD ! h2 rMZ= correlation between monozygotic co-twins rDZ= correlation between dizygotic co-twins • Assuming that shared environmental effects are equal for both types of twins. Twin Concordances Limitations of Twins !"#$%&'$($%) • Common Environment Unless Raised Apart • Twins reared apart are difficult to find • Common prenatal environment; 5)$6/ 1%2'7.82-"#'3 9'8.:3-*;'6.-4"5#.-"<'3 5;=; >3#'%*;'6.-4";"8-"66'.# >?1; !5; 1.%'3:*>2.@'3 AB; Variance Component Approach Modeling the Phenotype: p = µ + "# i xi + a + d+ e µ Population mean # Regression coefficients x Scaled covariates a Additive genetic effects d Dominance genetic effects e Random environmental effects – intrauterine competition • Mother may be physically stressed • Twin samples are fundamentally less powerful for QTL localization Variance Component Approach 2 2 2 $ = 2 %! a + & ! d + "! e 7 # coefficient of relationship between individuals $ probability that individuals share both alleles IBD " Kronecker function Defining Mendelian Relationships Relatives Parent-child Half siblings Full siblings Cousins Covariance 1/2VA 1/4VA 1/2VA+ 1/4VA 1/8VA Heritability b = 1/2 h2 r = 1/4 h2 r ! 1/2 h2 r = 1/8 h2 Kinship Matrix 2 4 3 5 Dad 1 ignoring interactions and shared environmental effects 1 Simple Kinship Matrix Mom 2 3 D M 1 2 3 D 1 0 % % % M 0 1 % % % 1 % % 1 % % 2 % % % 1 % 3 % % % % 1 Limitations of Heritability Estimates 1 2 3 4 5 1 1 0 0 % 0 2 0 1 0 % % 3 0 0 1 0 % 4 % % 0 1 & 5 0 % % & 1 1. Heritability is a population level parameter, summarizing the strength of genetic influences on variation in a trait among members of the population. It doesn’t tell you anything about particular individuals. 2. Heritability is an aggregate of the effects of multiple genes. It tells you nothing about how many genes influence a phenotype. A high heritability is not necessarily ‘better’ if it is due to many, many genes. Relative Risk Heritable vs. Familial The risk to a relative of an affected individual as compared to a randomly chosen member of the population, '. 1 - ( • A trait can be familial without being heritable. • Genes are not the only thing shared by members of a family (e.g. diet, exercise, environmental exposures, etc). • Nuclear families vs. extended pedigrees • Adoption studies Heritability of Volume & Gryi Bartely et al., Brain, 1997 10 MZ / 9 DZ pairs Gray-Matter Thickness Heritability Kremen et al., NeuroImage, 2010 110 MZ / 92 DZ pairs Vertex-Wide Cortical Thickness Winkler et al., Unpublished 726 family members Gray-Matter Thickness Heritability Thompson et al., Nat Neuro, 2001 10 MZ / 10 DZ pairs Cortical Thickness & Surface Area Winkler et al., NeuroImage, 2009 486 family members Superior Occipitofrontal Fascicle Hulshoff Pol et al. J. Neurosci, 2006 54 MZ / 58 DZ pairs / 34 Sibs White-Matter Tracts (DTI) h2 White-Matter Hyperintensity h2 Total Cranial: 0.91 Brain Parenchyma: 0.92 WMH: 0.73 Kochunov et al., NeuroImage, 2010 467 family members White-Matter Hyperintensity h2 74 MZ / 71 DZ pairs Task Based fMRI Heritability • Dorsal anterior cingulate cortex • Conflict Processing • h2=0.38 Whole-brain WMH: 0.72 Subcortical WMH: 0.66 Ependymal WMH: 0.73 Kochunov et al., Stroke, 2009 Carmelli et al., Stroke, 1998 459 family members Task Based fMRI Heritability Matthews et al., NeuroImage, 2007 10 MZ / 10 DZ pairs Resting State fMRI Heritability h2= 0.424 Koten et al., Science, 2009 10 MZ pairs / 10 sibs Glahn et al., Proc Nat Sci USA, 2010 333 family members Heritability Conclusions Question 2: Pleiotropy We’ve got heritability!! Which traits are influenced by the same genes? Levels of Pleiotropy Genetic Correlation (Pleiotropy) No Pleiotropy Partial Pleiotropy Full Pleiotropy Trait 1 Trait 2 Trait 1 Trait 2 Trait 1 Trait 2 Estimating Pleiotropy • Genetic correlation ()g): a measure of the overlap in genetic effects between traits. • )g varies from -1 to 1 • 0 = no pleiotropy; -1 or 1 = complete pleiotropy Cortical Thickness & Surface Area )p = )g'(h21h22) +)e'((1-h21)(1-h22)) h21 and h22 are the heritabilities of traits 1 and 2 Schmitt et al., Cerebral Cortex, 2008 107 MZ / 47 DZ / 228 others Winkler et al., NeuroImage, 2009 486 family members White-Matter Hyperintensity & Cortical Thinning & IQ WMH - Executive Function )g = 0.70 WMH - Mini-Mental State Exam )g = 1.0 )g = 1.0/1.0 Carmelli et al., Neurobiol Ageing, 2003 72 MZ / 70 DZ pairs White Matter Tracts & Superior longitudinal fasciculus – Spatial DRT: )g = 0.593 Karlsgodt et al., J Neurosci, 2010 Coffee Break 467 family members Brans et al., J Neurosci, 2010 )g = 1.0/0.7 77 MZ / 84 DZ pairs Pleiotropy Conclusions