* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture 7: Tetrad analysis
Survey
Document related concepts
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome evolution wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic drift wikipedia , lookup
Population genetics wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Microevolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Transcript
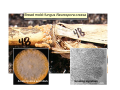
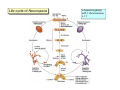
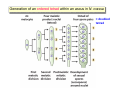
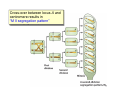
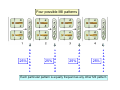
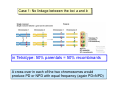
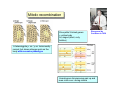
Lecture 7: Tetrad analysis 1. Life cycle of Neurospora 2. Gene to centromere mapping 3. Mapping the distance between two genes 4. Mitotic segregation and recombination Bread mold: fungus Neurospora crassa A colony on a petri dish Growing mycelium Life cycle of Neurospora a haploid organism with 7 chromosomes n=7 Generation of an ordered tetrad within an ascus in N. crassa = doubled tetrad Segregation of alleles during meiosis in Neurospora crassa: “M I segregation pattern” = doubled tetrad Two possible types of M I segregation pattern a a a a a A A A A A 50% 50% Cross-over between locus A and centromere results in “M II segregation pattern” Four possible MII patterns 25% A A a a A A a a 25% a a A A a a A A 25% A A a a a a A A 25% a a A A A A a a Each particular pattern is equally frequent as any other MII pattern Frequency of asci with MII patterns is used to determine map distance between the locus A and the centromere RF (A, CM) = ½ * MII asci / Total asci Why to multiply by ½ ? ...because in any MII ascus only ½ of the spores are recombinant Two simple formulas to remember RF (A, CM) = ½ * MII asci / Total asci RF max (A, CM) = 33% Gene-to-gene mapping: only three types of tetrads are possible for two pairs of alleles (regardless of linkage) Case 1: No linkage between the loci a and b PD=NPD Case 1: No linkage between the loci a and b in Tetratype: 50% parentals = 50% recombinants A cross-over in each of the two chromosomes would produce PD or NPD with equal frequency (again PD=NPD) Case 1: No linkage between the loci a and b - summary PD=NPD total parentals = total recombinants (RF = 50%) Case 2: the loci a and b are linked RF (a,b) = = recombinants total NPD + ½ T = Total asci PD>>NPD additional types of DCO result in PD or T and, therefore, remain unnoticed Mitotic non-disjunction: weird stuff M dominant allele for slender bristles in Drosophila M/M+, slender bristles M+/M+, normal bristles First described by C. Bridges in 1930-ies Mitotic non-disjunction: weird stuff M dominant allele for slender bristles in Drosophila M/M+, slender bristles M+/M+, normal bristles a a A A A/A/a a mitotic non-disjunction may reveal recessive alleles Mitotic recombination Drosophila X-linked genes: y, yellow body sn, singed (short, curly bristles) Discovered by Curt Stern in 1936 A heterozygote y+ sn / y sn+ looks mostly normal, but shows strange spots on the body with recessive phenotype Homologous chromosomes pair up and even cross-over during mitosis