* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Discovery of Entry Inhibitors for HIV-1: Predictions via a Novel De Novo Protein Design Framework and Experimental Validation
CCR5 receptor antagonist wikipedia , lookup
Discovery and development of HIV-protease inhibitors wikipedia , lookup
Drug discovery wikipedia , lookup
Discovery and development of antiandrogens wikipedia , lookup
NK1 receptor antagonist wikipedia , lookup
DNA-encoded chemical library wikipedia , lookup
Metalloprotease inhibitor wikipedia , lookup
Discovery and development of non-nucleoside reverse-transcriptase inhibitors wikipedia , lookup
Discovery and development of direct Xa inhibitors wikipedia , lookup
Discovery and development of neuraminidase inhibitors wikipedia , lookup
Drug design wikipedia , lookup
Discovery and development of integrase inhibitors wikipedia , lookup
Discovery and development of ACE inhibitors wikipedia , lookup
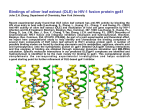
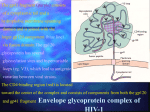
Discovery of Entry Inhibitors for HIV-1: Predictions via a Novel De Novo Protein Design Framework and Experimental Validation Meghan L. Bellows*, Hoki Fung, and Christodoulos A. Floudas Department of Chemical Engineering, Princeton University Lin Shen and Robert F. Siliciano Department of Medicine, Johns Hopkins University Martin S. Taylor and Phillip A. Cole Department of Pharmacology, Johns Hopkins University Session 3, Talk 2 (2:00 PM) A new de novo design framework with a ranking metric based on approximate binding affinity calculations is introduced. The framework consists of two stages: a sequence selection stage and a validation stage. The sequence selection stage produces a rank-ordered list of amino acid sequences with the lowest energies by solving an integer programming sequence selection model [1]. The validation stage uses both fold specificity calculations and approximate binding affinity calculations to re-rank the sequences from stage one, validating the sequence’s fold and binding to a target protein [2]. We applied the framework to develop short peptide-based inhibitors of HIV-1 entry into host cells. These peptidic inhibitors consist of 12 amino acids and target the core hydrophobic pocket of gp41. The choice of the epitope is based on: (i) the success of enfuvirtide (Fuzeon), (ii) the work on short constrained C-peptides that inhibit HIV-1 gp41, and (iii) the crystal structure elucidated on the best inhibitor complexed with the gp41 hydrophobic core [3]. A number of the best-predicted sequences were synthesized and their inhibition of HIV-1 was tested in vitro. All of the peptides examined showed inhibitory activity when compared with no drug present, and the novel peptide sequences outperformed the native template sequence used for the design. The best sequence showed micromolar inhibition. In addition, all peptides were shown to be non-toxic, maintaining their full viability at 500 μM. [1] [2] [3] H. K. Fung, M. S. Taylor, and C. A. Floudas, Optim. Methods & Software, 22 (2007). M. L. Bellows, H. K. Fung, C. A. Floudas, A. Lopez de Victoria, and D. Morikis, Biophys. J., submitted (2009). S. K. Sia, P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim, PNAS, 99 (2002).