* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Mayo Clinic Grand Rounds
Survey
Document related concepts
Transcript
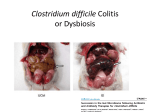
The Skin Microbiome Yoon K. Cohen, D.O. Hot Spots in Dermatology August 18, 2013 Objectives Human Microbiome Project Introduction of Skin Microbiome Factors Contributing to Variation in the Skin Microbiome Topographical Distribution of Microbes Microbes Commonly Found on Skin The Skin Microbiome and Diseases Human Microbiome Project 250 Healthy Volunteers 5 Sites Nasal passage Oral cavities GI Urogenital tract Skin 16S ribosomal RNA genomic sequening on 11,174 samples Conserved regions a binding site for PCR primer Hypervariable regions taxonomic classification Chen YE, et al. The skin microbiome: Current perspectives and futures challenges. J Am Acad Dematol. 2013; 69:143-155 Skin Microbiome Microbiology and dermatology have been intimately related The cutaneous surface is inhabited by myriad bacteria, fungi, and viruses. Now we begin to understand how these microbial communities impact human health and disease The skin microbiota plays a vital role in educating the immune system as the cutaneous innate and adaptive immune reponses can modulate the skin microbiota Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 Microbiome and skin immunology Chen YE, et al. The skin microbiome: Current perspectives and futures challenges. J Am Acad Dematol. 2013; 69:143-155 Skin Microbiome Three major questions What microbes are present on the skin surface? How does microbial diversity contribute to health and disease states? How do dermatologic practices alter microbial diversity? Kong HH, Segre JA. Skin Microbiome: Looking Back to Move Forward. J Invest Dermatol. 2011;132:933-39 Factors Contributing to Variation in the Skin Microbiome Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 Topographical Distribution of Bacteria on Skin Sites Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 Composition of a Single Metagenome Chen YE, et al. The skin microbiome: Current perspectives and futures challenges. J Am Acad Dematol. 2013; 69:143-155 Interpersonal Variation of the Skin Microbiome This chart demonstrates that skin microbial variation is more dependent on the site than on the individual. Bars represents the relative abundance of bacterial taxa as determined by 16S ribosomal RNA sequencing Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 Temporal diversity of the microbiome Oh et al, Shifts in human skin and nares microbiota of healthy children and adults. Genome Medicine. 2012 Bacteria Commonly Found on Skin Staphylococcus epidermidis Staphylococcus aureus Corynebacterium spp. Propionibacterium acnes S. Epidermidis Pathogen Frequent cause of nosocomial infections Immunocomprised patients Indwelling devices Commensal Major skin inhabitant Produce antibacterial products Bacteriocins (epidermin, epilancin K7, Pep5, staphlococcin 1580) A clump of Staphylococcus epidermidis bacteria S. Aureus Pathogen Frequent cause of infections (self-limited to invasive) Methicillin-resistance is a healthcare problem Phenol-soluble modulins (PSMs) produced in high levels by CA-MRSA Streptococcus pyogenes is sensitive to S. aureus PSMs, which may partically explain CA-MRSA dominance Commensal Asymptomatic nasal colonizers 20% permanently colonized 30-50% transiently colonized S. aureus preferentially hemolyzes human blood to utilizes iron from heme to promote proliferation Can produce bacteriocin (staphylococcin 462) Corynebacterium spp. Pathogen Diphtheroids +/- C. dephtheriae Part of normal skin flora C. minutissimum (erytherasma) and C. tenuis (trichomycosis) Risk factors for infections Immunocompromised patients Skin barrier defects Commensal Prevents oxidative damage by producing superoxide dismutase Produce bacteriocin-like compounds Propionibacterium acnes Pathogen Associated with folliculitis, systemic infections and acnes Commensal Produce bacteriocin-like compounds with activity against bacteria, yeast and molds Other Microbes Commonly Found on Skin Malassezia spp. Demodex mites Human papillomavirus (HPV) Identifying Fungi and Viruses Fungi Similar strategy can be used to classify the 18S rRNA or the intervening sequence (ITS) of fungi Viruses De-novo sequencing Challenging what to use for control for DNA or RNA viruses Currently resequencing the human genome to identify viral associated disease Once you find them, finding them again is PCR-based Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 Relative abundance of fungal genera and Malassezia species at different human skin sites. Fungal diversity of individual body sites of healthy volunteers (1–10) was taxonomically classified at the genus level, with further resolution of Malassezia species. Findley et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. June 2013 Skin Diseases Associated with Dysbiosis Gallo RL, Nakatsuji T. Microbial Symbiosis with the innate immune Defense System of the Skin. J invest Dermatol. 2011 Conclusions Skin Microbiome How molecular approaches allow us to better understand the relationship between skin microbiome and human health & disease states Currently active ongoing research for skin microbiome under NIH Human Microbiome Project Future Therapeutic Options The impact of repeated use of topical/systemic antimicrobial therapies Mainstay of dermatologic practice Associated risks are not fully understood Alternative therapies Probiotic microbial organisms Antimicrobial chemicals derived from microorganisms or humans References 1. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011; 9: 244-53 2. Kong HH, Segre JA. Skin Microbiome: Looking Back to Move Forward. J Invest Dermatol. 2012; 132: 933-39 3. Capone KA. Dowd SE, Stamatas GN, et al. Diversity of the Human Skin Microbiome Early in Life. J invest Dermatol. 2011; 131: 2026-32 4. Gallo RL, Nakatsuji T. Microbial Symbiosis with the innate immune Defense System of the Skin. J invest Dermatol. 2011; 131: 1974-80 5. Gaspari AA, et al. Chapter 9. “Antimicrobial Peptides”. Clinical and Basic Immunodermatology. Springer. London. 2009 6. Zimmer C. Tending the Body’s Microbial Garden. The New York Times. June 18, 2012 7. Specter M. Germs Are Us. The New Yorker. October 22, 2012 8. Gorman C. Explore the Human Microbiome. Scientific American. May 15, 2012 The Sebago Lake in Maine, May 27th, 2012